Home > Organization > Divisions and Independent Research Units > Division of Rare Cancer > Research Projects > Establishment of fundamental research resource
Establishment of fundamental research resource
Rare cancer research has a unique problem; “the number of patients is small, and the clinical materials are hardly available for research”. Probably because the clinical materials are not well available for research, the fundamental research tools are not well developed for the rare cancer research. Division of Rare Cancer Research addresses this issue.
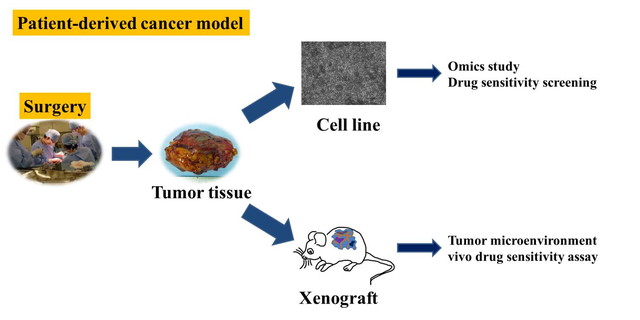
Establishment of patient-derived cancer model
Patient-derived xenografts (PDXs) and patient-derived cells (PDCs) mimic the in vivo environment, and have been considered as essential tools for cancer research. The experiments using PDXs and PDCs can generate valuable information, which cannot be available otherwise. For example, the functional significances of novel genetic aberrations, and the mode of action of new anti-cancer drugs should be revealed by the experiments using living cancer cells. The PDXs and PDCs can be shared in the research community, facilitating cancer research. Patient-derived cancer models have been developed in many types of malignancies. However, they are not available in rare cancers. In the rare cancers, even cell lines are not well available. For example, in sarcomas, only a limited number of cell lines are publicly available from the cell banks. Considering that there are more than 50 histologically different sarcomas, and the treatments largely depend on the histology, patient-derived cancer models for individual sarcomas are an urgent requirement. Moreover, the cell lines available from the public cell banks have several limitations in quality; the information of clinical and genetic backgrounds of cell lines are not opened, and the diagnostic criteria for the original tumors of cell lines are out of data. These problems are not limited to sarcomas, but also exist in other rare cancers.
Division of Rare Cancer addresses these problems by collaborating with Department of Innovative Seeds Evaluation and Central Animal Division in the institute, Division of Musculoskeletal Oncology in the hospital. We have been focusing on sarcomas, and established patient-derived sarcoma models using surgically resected tumor tissues. The established models are pathologically examined in collaboration with Department of Pathology and Clinical Laboratories in the hospital. The molecular backgrounds of the established models are examined to determine the genetic aberrations, which can be targets of molecular targeting drugs. We also collaborate with Division of Brain Tumor Translational Research in the institute to establish the models of malignant brain tumors. The procedures to establish the in vitro models depend on the histology of malignancies, and we try to establish the protocols to improve the efficacy of establishment. We also aim to construct the system to share the established models with other researchers.
We launched the project of patient-derived cancer model in 2014. Since then, we established many patient-derived cancer models. We use them to develop the novel therapy for rare cancers.
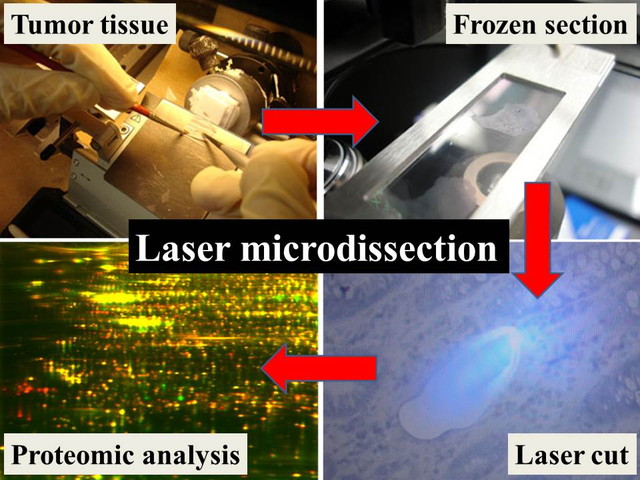
Development of experiment technology
The clinical materials for research are generally limited in number and amount. Especially in rare cancer, this issue influences the research considerably. It is quite important concept to transform the limited research resources such as tumor tissues to the unlimited ones. For this sake, the establishment of patient-derived cancer model is an effective approach. Bioinformatics can also transform the limited resources to the unlimited ones by using “information” which can be replicated and modified unlimitedly. In addition, we developed the proteomics modalities which allow us to investigate the proteome using a very limited amount of tissue samples. Our method enables proteomic study using only 1000-3000 tumor cells, and it has been used by many domestic and oversea researchers. The technology to use a limited amount of clinical samples will be useful not only in the rare cancer studies, but also in all cancer researches.
Links.
- References 1 : Application of highly sensitive fluorescent dyes (CyDye DIGE Fluor saturation dyes) to laser microdissection and two-dimensional difference gel electrophoresis (2D-DIGE) for cancer proteomics. (Link to External Site.)
Bioinformatics approach
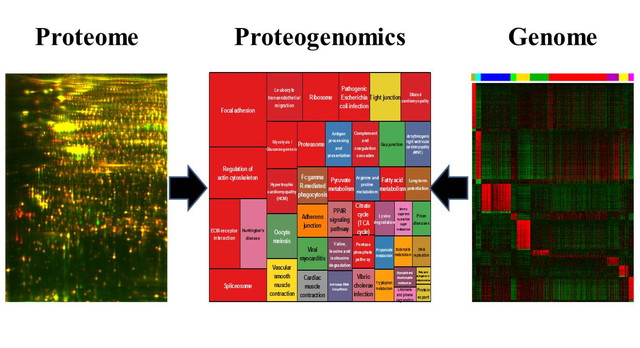
We can obtain the information about the events in our body by the experiments using clinical materials. The experiments using patient-derived cancer models are quite important for the translational research. However, the results and hypothesis should be validated by the experiments using clinical materials. Omics studies have been conducted in many types of malignancies, and the genetic aberrations relevant to metastasis, recurrence and resistance to treatments were reported. The data were deposited to the public database, and used in other studies. In rare cancers, the database can be improved in terms of construction and application.
We conduct the study on global genetic aberrations of rare cancers, and accumulate the data in our original rare cancer database. Our database will be opened and browsed on the websites. The database, which allows integrate the omics data with the clinical pathological information will promote the rare cancer research. For example, we constructed proteome database (References 1 and 2). Our proteome database, namely Genome Medicine Database of Japan (GeMDBJ Proteomics), was accessed by more than 600,000 times. As a database of disease proteomics, GeMBJ Proteomics is a unique research resource. Currently, we are constructing the database of transcriptome and genome of rare cancers.
The omics data of rare cancers have been generated using a small number of samples in many laboratories. Parts of the results are opened on line. Although the number of cases in individual studies is small, as a total, a considerable number of cases were subjected to the omics studies. We conduct meta-analysis of those data by integrating the data from academic papers. As the clinical materials for research are limited in the rare cancers, the use of existing omics data is especially required. The organization of clinical information of individual omics data sets and the application of bioinformatics are quite important for such studies. In Division of Rare Cancer Research, the researches with medical backgrounds curate the clinical and pathological information of cases for omics studies by reading academic papers, and register the results to our custom database. The data from in vitro cultured cell lines are also integrated to our analysis. We aim to develop the methodology to identify the innovative seeds for novel therapy.
In addition to biomarker development, the bioinformatics is useful for the study of re-localization of existing anti-cancer drugs to rare cancers. Using the aforementioned original database, we are developing the programs to identify the anti-cancer drugs for rare cancers in collaboration with Opt (http://www.opt.ne.jp/en/).
Links.
- Reference 1 : Cancer proteome-expression database: Genome Medicine Database of Japan Proteomics. (Link to External Site.)
- Reference 2 : Proteome Expression Database of Ewing Sarcoma: a Segment of the Genome Medicine Database of Japan Proteomics (Link to External Site.)
- Opt Inc. (Link to External Site.)
Multi-institutional collaboration and networking
We conduct the multi-institutional collaborations to investigate the novel innovative seeds for rare cancers by using clinical materials. The participants include the domestic universities which conduct clinical practice and research of rare cancers, as well as the oversea researchers who focus on the rare cancers. In addition to the establishment of patient-derived cancer models of sarcomas and malignant brain tumors, we perform the multi-layer omics studies of rare cancers. Our collaborations will enable the research which is difficult to be done by single institutions. We will share the research outcomes by discussions, so that all participants can be benefited by the collaborations. We will use the results of collaborations to promote novel therapies by getting patents and industrialization of the developed innovative seeds.

