HOME > Publication & Reports > Annual Report 2016 > Exploratory Oncology Research & Clinical Trial Center
Division of Translational Genomics (Kashiwa Campus)
Katsuya Tsuchihara, Hideki Makinoshima, Sachiyo Mimaki, Shingo Matsumoto, Wataru Okamoto, Ayako Suzuki, Atsushi Yagishita, Rumi Fujioka, Hiroko Kumakura, Megumi Iwakura, Sayuri Todoroki, Hitomi Okazaki, Ayame Takeda, Tomohiro Sakamoto, Hiroyasu Esumi, Yutaka Suzuki, Shin Kawano, Tatsunosuke Ikemura, Keita Masuzawa, Satoshi Tada, Chikako Nakai
Introduction
The Division of Translational Genomics closely collaborates with intramural and extramural basic and clinical researchers to develop novel anti-cancer therapeutics as well as to prove their concepts. Our division has been developing genome biomarker diagnostics, exploring rational molecular targets for anti-cancer therapies, and elucidating molecular mechanisms of oncogenesis, tumor progression, and therapeutic responses. Our division is managing a data center called SCRUM-Japan, a nation-wide cancer genome screening project for developing new anti-cancer drugs.
Our team and what we do
Weekly conferences for the whole division and individual research groups are held. As a SCRUM-Japan steering committee member, our division contributed to quarterly held committee meetings and ad hoc database working group meetings.
Research activities
1)Identification of driver gene alterations of pulmonary large-cell neuroendocrine carcinoma has been done. As well as small cell lung cancer genomes that our division published, activating mutations of PI3K pathway molecules were detected.
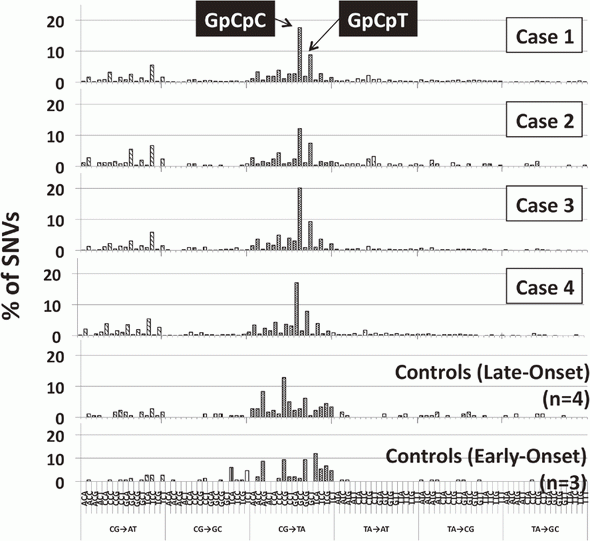
2)Whole exome analysis of occupational cholangiocarcinoma cases has been done. These cases, exposed to high-dose halokanes, showed hypermutation and unique mutational signatures (Figure 1).
3)The first selective fluorescent probe for class 3 ALDH (ALDH3A1) was developed. The probe will be useful for elucidating carcinogenesis of esophageal cancer.
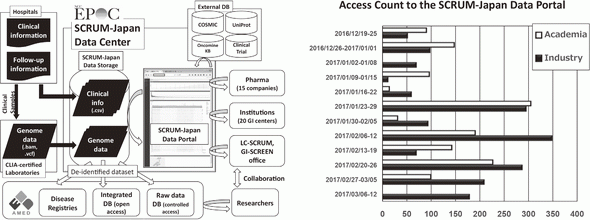
4)SCRUM-Japan data center has been integrating clinical and genome analysis data of more than 7,500 cases and distributing these data to the researchers with the collaboration of academic institutes and pharmaceutical companies (Figure 2).
Clinical trials
1)Screening project for Individualized Medicine in Japan (SCRUM-Japan): Data center
Education
Our division accepted and trained the following trainees: Graduate students from the University of Tokyo and Keio University, staff physicians and residents of the National Cancer Center Hospital East, Tottori University and Keio University, visiting scientists from the Tokyo University of Science, the University of Tokyo, Tottori University, Keio University, diagnostics and IT companies.
Future prospects
SCRUM-Japan is establishing its reputation for a role model of cancer precision medicine. Our division will feed back the know-how to propel the clinical application of genome-based cancer precision medicine.
List of papers published in 2016
Journal
1.Shiraishi K, Okada Y, Takahashi A, Kamatani Y, Momozawa Y, Ashikawa K, Kunitoh H, Matsumoto S, Takano A, Shimizu K, Goto A, Tsuta K, Watanabe S, Ohe Y, Watanabe Y, Goto Y, Nokihara H, Furuta K, Yoshida A, Goto K, Hishida T, Tsuboi M, Tsuchihara K, Miyagi Y, Nakayama H, Yokose T, Tanaka K, Nagashima T, Ohtaki Y, Maeda D, Imai K, Minamiya Y, Sakamoto H, Saito A, Shimada Y, Sunami K, Saito M, Inazawa J, Nakamura Y, Yoshida T, Yokota J, Matsuda F, Matsuo K, Daigo Y, Kubo M, Kohno T. Association of variations in HLA class II and other loci with susceptibility to EGFR-mutated lung adenocarcinoma. Nat Commun, 7:12451, 2016
2.Ikeda M, Sato A, Mochizuki N, Toyosaki K, Miyoshi C, Fujioka R, Mitsunaga S, Ohno I, Hashimoto Y, Takahashi H, Hasegawa H, Nomura S, Takahashi R, Yomoda S, Tsuchihara K, Kishino S, Esumi H. Phase I trial of GBS-01 for advanced pancreatic cancer refractory to gemcitabine. Cancer Sci, 107:1818-1824, 2016
3.Mimaki S, Totsuka Y, Suzuki Y, Nakai C, Goto M, Kojima M, Arakawa H, Takemura S, Tanaka S, Marubashi S, Kinoshita M, Matsuda T, Shibata T, Nakagama H, Ochiai A, Kubo S, Nakamori S, Esumi H, Tsuchihara K. Hypermutation and unique mutational signatures of occupational cholangiocarcinoma in printing workers exposed to haloalkanes. Carcinogenesis, 37:817-826, 2016