HOME > Publication & Reports > Annual Report 2016 > Exploratory Oncology Research & Clinical Trial Center
Division of Translational Genomics (Tsukiji Campus)
Takashi Kohno, Hitoshi Ichikawa, Takashi Kubo
Introduction
This division aims to facilitate precision cancer medicine by developing a next-generation sequencing (NGS)-based genomic testing system and advancing its clinical implementation.
Research activities
1.Establishment of an NGS-based genomic testing system
We developed an NGS-based in-house genomic testing system to identify somatic gene mutations, amplifications, and fusions using an original cancer gene panel (NCC oncopanel), and have been continuously improving this system to become a clinically useful in vitro diagnostics (IVD) system in cooperation with the Department of Clinical Genomics and the Department of Bioinformatics of the National Cancer Center Research Institute (NCCRI). This system was improved as follows. (1) The gene panel was renewed as NCC oncopanel v4. (2) Paired analysis of tumor and normal tissue DNAs were performed to distinguish somatic mutations and germline mutations. (3) A new data analysis pipeline was developed to detect known complex mutations in EGFR and others, which could not be detected in ordinary NGS analysis. (4) Reporting system was renewed by constructing a knowledge base including well-known druggable gene aberrations. In cooperation with staff of the Department of Pathology and Clinical Laboratories in the National Cancer Center Hospital (NCCH), the SCI-Lab was opened in the NCCH, where our clinical sequencing system is operated under an international standard quality assurance.
2.Implementation of NGS-based genomic testing in the NCCH
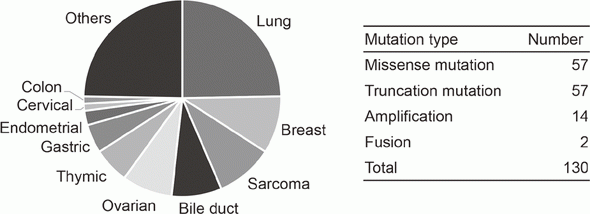
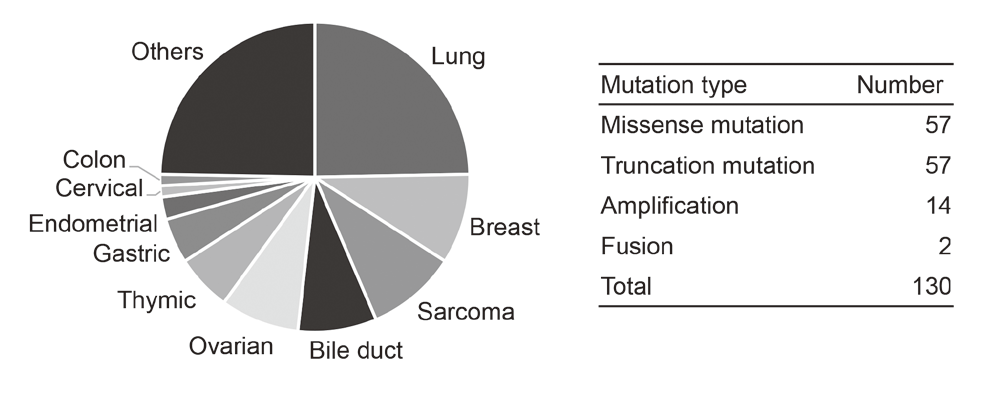
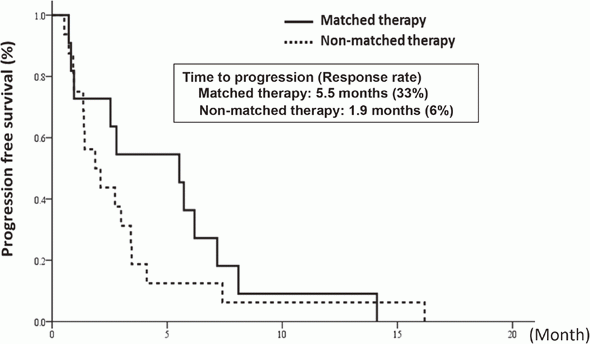
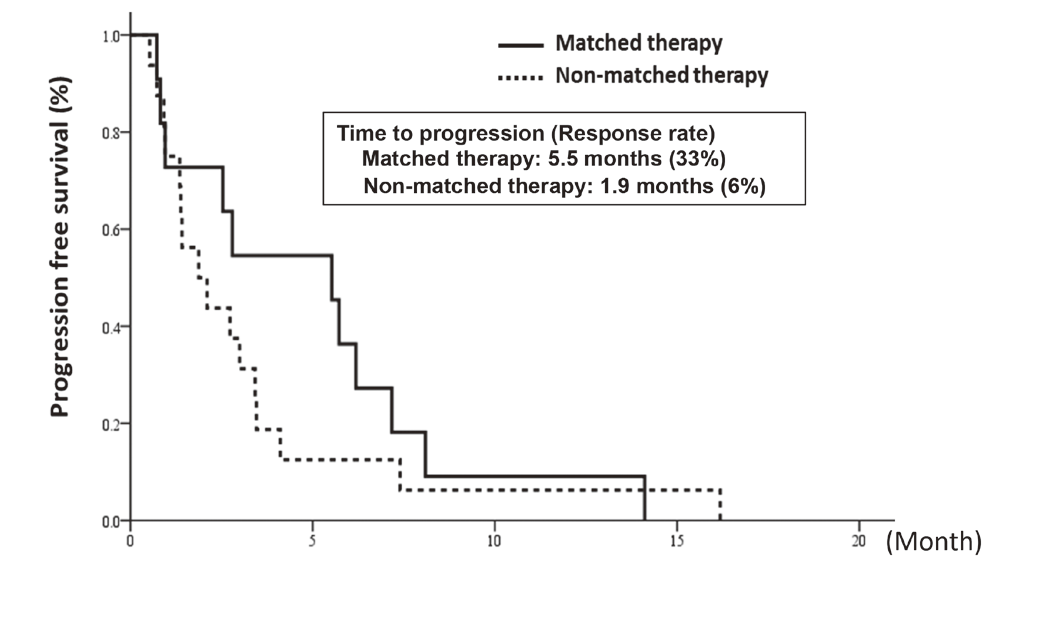
In cooperation with staff of the Department of Experimental Therapeutics and the Department of Pathology and Clinical Laboratories in the NCCH, prospective clinical studies were performed to examine the feasibility and utility of our NGS-based genomic testing system for patients enrolled into early-phase clinical trials. The result of the TOPICS-1 (TOP-GEAR-1) study, in which patients were enrolled in 2013 and 2014, indicated that patients who received investigational drug therapies matched to their genomic aberrations showed better progression-free survivals than those who received non-matched therapies (Figure 1). In 2016, 85 patients were analyzed in the TOP-GEAR-1ex study in which NGS testing was performed in the NCCH (Figure 2), and additional 94 patients were analyzed in the TOP-GEAR-2 study in which NGS testing was performed in the SCI-Lab. In the TOP-GEAR-2 study, actionable genomic aberrations were identified in 64 (68%) patients. Germline mutations in genes responsible for hereditary cancers were identified in two patients as a secondary finding. Diagnosis, treatment, and care for patients with germline mutations are undertaken by staff of the Department of Genetic Medicine and Services (GeMS) in the NCCH.
Clinical trials
TOP-GEAR: Trial of Onco-Panel for Gene-profiling to Estimate both Adverse events and Response by cancer treatment (UMIN000011141)
Education
Post-doctoral fellows and chief residents in NCC were educated through the "on-the-job training" in several translational research projects.
Future prospects
The feasibility and clinical utility of our genomic testing system with international standard quality assurance will be shown. A clinical study for approval of our system by the Pharmaceuticals and Medical Devices Agency (PMDA) will be launched shortly.
Figure 1. Progression-free survival of matched and non-matched therapy-received patients in the TOP-GEAR-1 study
List of papers published in 2016
Journal
1.Seki Y, Fujiwara Y, Kohno T, Takai E, Sunami K, Goto Y, Horinouchi H, Kanda S, Nokihara H, Watanabe S, Ichikawa H, Yamamoto N, Kuwano K, Ohe Y. Picoliter-Droplet Digital Polymerase Chain Reaction-Based Analysis of Cell-Free Plasma DNA to Assess EGFR Mutations in Lung Adenocarcinoma That Confer Resistance to Tyrosine-Kinase Inhibitors. Oncologist, 21:156-164, 2016
2.Kamata T, Sunami K, Yoshida A, Shiraishi K, Furuta K, Shimada Y, Katai H, Watanabe S, Asamura H, Kohno T, Tsuta K. Frequent BRAF or EGFR mutations in ciliated muconodular papillary tumors of the lung. J Thorac Oncol, 11:261-265, 2016
3.Sunami K, Furuta K, Tsuta K, Sasada S, Izumo T, Nakaoku T, Shimada Y, Saito M, Nokihara H, Watanabe S, Ohe Y, Kohno T. Multiplex Diagnosis of Oncogenic Fusion and MET Exon Skipping by Molecular Counting Using Formalin-Fixed Paraffin Embedded Lung Adenocarcinoma Tissues. J Thorac Oncol, 11:203-212, 2016
4.Asao T, Fujiwara Y, Sunami K, Kitahara S, Goto Y, Kanda S, Horinouchi H, Nokihara H, Yamamoto N, Ichikawa H, Kohno T, Tsuta K, Watanabe S, Takahashi K, Ohe Y. Medical treatment involving investigational drugs and genetic profile of thymic carcinoma. Lung Cancer, 93:77-81, 2016
5.Tanabe Y, Ichikawa H, Kohno T, Yoshida H, Kubo T, Kato M, Iwasa S, Ochiai A, Yamamoto N, Fujiwara Y, Tamura K. Comprehensive screening of target molecules by next-generation sequencing in patients with malignant solid tumors: guiding entry into phase I clinical trials. Mol Cancer, 15:73, 2016