HOME > Publication & Reports > Annual Report 2016 > Research Institute
Division of Epigenomics
Toshikazu Ushijima, Satoshi Yamashita, Hideyuki Takeshima, Naoko Hattori, Masahiro Maeda, Emi Kubo, Naoko Iida, Zong Liang, Mika Wakabayashi, Akiko Mori, Kana Kimura, Kanako Sakashita, Yuko Miyaji, Naoko Kobayashi, Aya Nakajima
Introduction
The Division of Epigenomics has been focusing on the epigenetic mechanisms of carcinogenesis, and has identified many aberrantly methylated genes in various cancers, including gastric cancers, esophageal squamous cell carcinomas (ESCCs), neuroblastomas, breast cancers, pancreatic cancers, lung cancers, ovarian cancers, and melanomas. These findings have led to the identification of novel tumor-suppressor genes, the development of powerful biomarkers, and the establishment of the concept of "epigenetic field for cancerization (field defect)". This division continues its activity in 1) developing clinically useful biomarkers, a novel approach to cancer prevention, and epigenetic therapy, and 2) revealing molecular mechanisms of aberrant DNA methylation induction.
Research activities
1.Identification of Novel Epigenetic Alterations
Identification of cancer-related pathways activated or inactivated by epigenetic alterations, such as aberrant DNA methylation, is important. This year, an integrated analysis of genetic and epigenetic alterations in ESCCs was conducted using next-generation sequencing and BeadArray technologies. It was revealed that the p53 signaling pathway and chromatin remodeling factors were inactivated mainly by genetic alterations in ESCCs. In contrast, the WNT signaling pathway was activated by epigenetic alterations, namely aberrant DNA methylation, of its negative regulators.
2.Development of Biomarkers
This division previously revealed that the degree of accumulated aberrant DNA methylation in normal-appearing gastric mucosae is expected to be a useful diagnostic marker to predict gastric cancer risk. To bring this concept into clinical practice, a multicenter prospective cohort study has been conducted for the prediction of metachronous gastric cancer risk after endoscopic resection. This year, a final analysis was conducted, and the concept "the measurement of aberrant DNA methylation accumulated in normal tissues is clinically useful for cancer risk diagnosis", which we showed by an intermediate analysis of this study, was established. Based on these results, we started a multicenter prospective cohort study for the prediction of gastric cancer risk in healthy volunteers who underwent eradication of Helicobacter pylori (H. pylori), the almost exclusive cause of gastric cancers. This year, an initial methylation profile of healthy volunteers was determined.
Contamination of normal cells is almost always present in tumor samples and affects their molecular analyses, and development of a biomarker that can estimate the fraction of cancer cells in a tumor DNA sample is important. This year, we identified three genomic regions, OSR2, PPFIA3, and VAV3, whose DNA methylation levels reflect the fraction of cancer cells in gastric tumor DNA samples.
3.Development of epigenetic cancer prevention
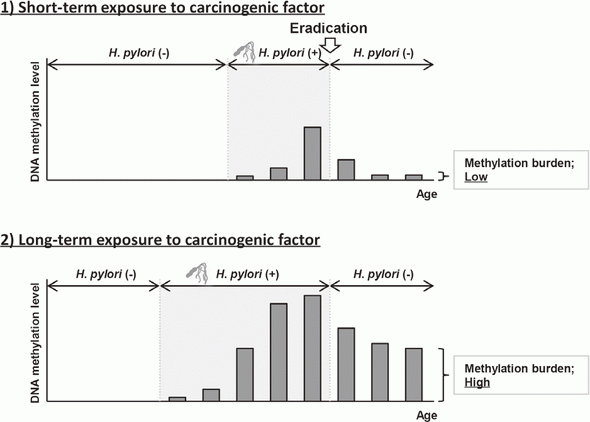
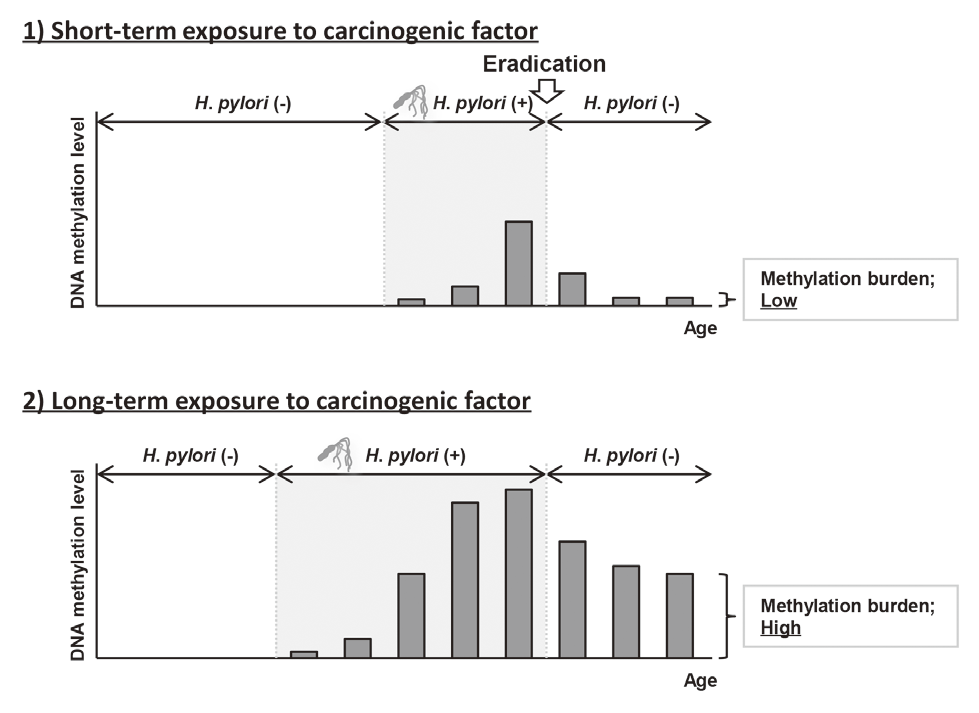
Elimination of aberrant DNA methylation accumulated in normal tissues, namely methylation burden, is a novel approach to cancer prevention that has the potential to reduce cancer risk once accumulated. This year, using a Mongolian gerbil model of H. pylori-induced gastritis, it was revealed that the levels of methylation burden in normal tissues are determined by the exposure period to a carcinogenic factor (Figure 1).
This suggested that the elimination of exposure to carcinogenic factors, such as H. pylori eradication in the stomach, in its early stage is beneficial for cancer prevention.
Future prospects
Based on these results, this division will 1) continue multicenter prospective cohort studies for the prediction of gastric cancer risk, 2) conduct the development of epigenetic cancer prevention and therapy, and 3) reveal the molecular mechanisms of how aberrant DNA methylation is induced by chronic inflammation.
Other activities
This division assisted with 1) epigenetic and genetic analyses of primary cancer samples in several translational researches conducted in the National Cancer Center and other institutions, and 2) epigenetic analysis in various animal models.
List of papers published in 2016
Journal
1.Matsuda Y, Ishiwata T, Yoshimura H, Yamashita S, Ushijima T, Arai T. Systemic administration of small interfering RNA targeting human nestin inhibits pancreatic cancer cell proliferation and metastasis. Pancreas, 45:93-100, 2016
2.Kuboki Y, Yamashita S, Niwa T, Ushijima T, Nagatsuma A, Kuwata T, Yoshino T, Doi T, Ochiai A, Ohtsu A. Comprehensive analyses using next-generation sequencing and immunohistochemistry enable precise treatment in advanced gastric cancer. Ann Oncol, 27:127-133, 2016
3.Hattori N, Ushijima T. Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med, 8:10, 2016
4.Nakazato H, Takeshima H, Kishino T, Kubo E, Hattori N, Nakajima T, Yamashita S, Igaki H, Tachimori Y, Kuniyoshi Y, Ushijima T. Early-stage induction of SWI/SNF mutations during esophageal squamous cell carcinogenesis. PLoS One, 11:e0147372, 2016
5.Zong L, Hattori N, Yoda Y, Yamashita S, Takeshima H, Takahashi T, Maeda M, Katai H, Nanjo S, Ando T, Seto Y, Ushijima T. Establishment of a DNA methylation marker to evaluate cancer cell fraction in gastric cancer. Gastric Cancer, 19:361-369, 2016
6.Sasaki Y, Hamaguchi T, Yamada Y, Takahashi N, Shoji H, Honma Y, Iwasa S, Okita N, Takashima A, Kato K, Nagai Y, Taniguchi H, Boku N, Ushijima T, Shimada Y. Value of KRAS, BRAF, and PIK3CA Mutations and Survival Benefit from Systemic Chemotherapy in Colorectal Peritoneal Carcinomatosis. Asian Pac J Cancer Prev, 17:539-543, 2016
7.Ichimura K, Fukushima S, Totoki Y, Matsushita Y, Otsuka A, Tomiyama A, Niwa T, Takami H, Nakamura T, Suzuki T, Fukuoka K, Yanagisawa T, Mishima K, Nakazato Y, Hosoda F, Narita Y, Shibui S, Yoshida A, Mukasa A, Saito N, Kumabe T, Kanamori M, Tominaga T, Kobayashi K, Shimizu S, Nagane M, Iuchi T, Mizoguchi M, Yoshimoto K, Tamura K, Maehara T, Sugiyama K, Nakada M, Sakai K, Kanemura Y, Nonaka M, Asai A, Yokogami K, Takeshima H, Kawahara N, Takayama T, Yao M, Kato M, Nakamura H, Hama N, Sakai R, Ushijima T, Matsutani M, Shibata T, Nishikawa R. Recurrent neomorphic mutations of MTOR in central nervous system and testicular germ cell tumors may be targeted for therapy. Acta Neuropathol, 131:889-901, 2016
8.Sekine S, Yamashita S, Tanabe T, Hashimoto T, Yoshida H, Taniguchi H, Kojima M, Shinmura K, Saito Y, Hiraoka N, Ushijima T, Ochiai A. Frequent PTPRK-RSPO3 fusions and RNF43 mutations in colorectal traditional serrated adenoma. J Pathol, 239:133-138, 2016
9.Abe M, Yamashita S, Mori Y, Abe T, Saijo H, Hoshi K, Ushijima T, Takato T. High-risk oral leukoplakia is associated with aberrant promoter methylation of multiple genes. BMC Cancer, 16:350, 2016
10.Mori G, Nakajima T, Asada K, Shimazu T, Yamamichi N, Maekita T, Yokoi C, Fujishiro M, Gotoda T, Ichinose M, Ushijima T, Oda I. Incidence of and risk factors for metachronous gastric cancer after endoscopic resection and successful Helicobacter pylori eradication: results of a large-scale, multicenter cohort study in Japan. Gastric Cancer, 19:911-918, 2016
11.Ushijima T, Yoshino T. The moment that KRAS mutation started to evolve into precision medicine in metastatic colorectal cancer. Cancer Res, 76:6443-6444, 2016
12.Kishino T, Niwa T, Yamashita S, Takahashi T, Nakazato H, Nakajima T, Igaki H, Tachimori Y, Suzuki Y, Ushijima T. Integrated analysis of DNA methylation and mutations in esophageal squamous cell carcinoma. Mol Carcinogenesis, 55:2077-2088, 2016