HOME > Publication & Reports > Annual Report 2016 > Research Institute
Division of Cancer Genomics
Tatsuhiro Shibata, Fumie Hosoda, Yasushi Totoki, Shinichi Yachida, Yasuhito Arai, Natsuko Hama, Hiromi Nakamura, Hirofumi Rokutan, Erina Takai, Koichi Ogura, Akihiro Ohmoto, Mihoko Adachi, Masami Suzuki, Hiroko Shimizu, Shoko Ohashi, Wakako Mukai, Erika Arakawa, Seri Yamagishi, Hiroe Nozaki, Machiko Watanabe, Risa Usui, Arisa Kaya, Kazuko Murakami
Introduction
The Division of Cancer Genomics focuses on the comprehensive characterization of cancer genome on the basis of tumor pathology, and aims to make a "breakthrough" by identifying novel cancer-related genes, including potential therapeutic targets, and biomarkers, and to understand the cancer genome as heterogeneous but intervention-able "biological systems" that contribute to the pathogenesis of cancer. This Division has also been participating in the international consortium (International Cancer Genome Consortium; ICGC), contributed to the core facility of the center, and developing new informatics tools for data analysis from various types of next-generation high-performance sequencers (NGS).
Research activities
1. Genomic analysis and translational research for intractable cancers
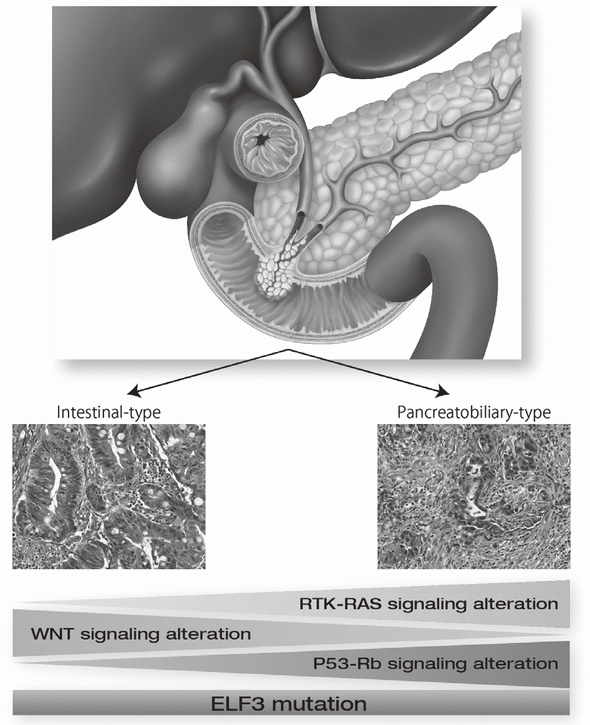
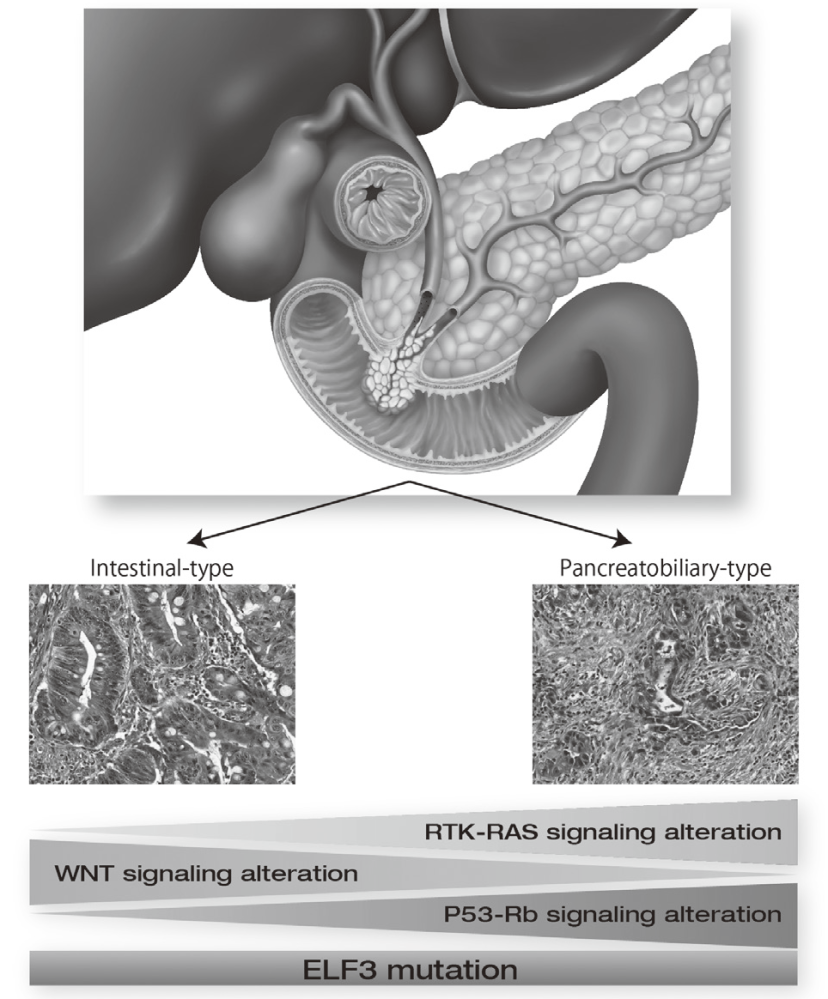
Exome and targeted sequencing in 172 ampullary carcinomas discovered ELF3 as a characteristic tumor suppressor gene and identified potential therapeutically targetable mutations in 51% of cases (Figure 1). Seven mutational signatures were identified in 266 liver cancer whole genomes, which associate with unique epidemiological backgrounds and somatic mutations status. In PRELUDE program (genomic screening consortium for biliary tract cancer) collaborated with the National Cancer Center Hospital (NCCH), molecular diagnosis for FGFR2 fusion genes in FFPE biopsy samples was established.
2. Genomic analysis and translational research for rare cancers
A novel fusion gene, NUTM2A-CIC, was discovered in undifferentiated round cell sarcoma cases. Capture sequencing of HTLV-1 genome in ATL carrier lymphocytes could precisely estimate virus copy number consistent with that detected by an ordinary quantitative PCR. Sequencing of HTLV-1 genome was also profitable to find structural alterations of the virus genome from HTLV-1 carrier blood directly. Exome and target sequencing of 68 mucinous gastric cancers, a rare subtype, identified a unique mutational profile and recurrent loss of function mutations in the MYH9 gene.
3. Genomic profiling of tumor environments (immune cells and microbiota)
Characterization of tumor-infiltrating lymphocytes, a direct effector of tumor immunity, is important for the development of cancer immunotherapy. A large-scale RNA sequencing of T-cell receptor beta chain was performed in gastric, colorectal and liver cancers including metastasis. Simple storage of faecal samples at room temperature, whose bacterial composition preserved as frozen ones, was established. We have characterized luminal microbiota in normal and colorectal cancer patients, and are exploring novel biological roles of microbiota in colorectal cancer development.
4. Cancer-associated germline genetics (familial cancer and pharmacogenomics)
Targeted sequencing of 21 pancreatic cancer susceptibility genes in Japanese familial pancreatic cancer patients identified eight (14.5%) patients carried deleterious variants in BRCA2, PALB2, ATM and MLH1. Target germline sequencing of 147 drug metabolism-related genes (NCC-PGx panel) in 75 ALK-positive lung cancer patients treated with crizotinib identified potential SNPs associate with severe adverse effects.
5. Development of analysis pipelines for whole genome/epigenome analysis
In-house pipeline for detecting genomic rearrangements (GR) including transposon mobilization was established. Comparison tools between GR breakpoints and nearby gene expression revealed potential enhancer-hijacked genes, including eight oncogenes, whose expressions were increased with GR of their regulatory regions. Furthermore, six insulator-disrupted genes were identified, and the top three of them were reported as cancer-related genes.
Education
Six young researchers, one cancer specialist training doctor, and one visiting researcher have been trained in this division. In addition, three young bioinformaticians have also been trained.
Future prospects
By utilizing current and cutting-edged sequencing technologies (e.g. long-read and single cell sequencing), this division will actively investigate the cancer genomics from both basic (new biomarkers including therapeutic targets, epigenomics, metagenomics and immune-genomics), and translational research (preclinical research, liquid clinical sequencing, PGx and germline evaluation) viewpoints. Especially tighter collaboration with cancer-immunology groups, by applying single cell immune-profiling and TCR repertoire sequencing will be achieved. This division will also contribute to the development of bioinformatics tools and human resources for analyzing the large cancer genomics data.
List of papers published in 2016
Journal
1.Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, Totoki Y, Fujimoto A, Nakagawa H, Shibata T, Campbell PJ, Vineis P, Phillips DH, Stratton MR. Mutational signatures associated with tobacco smoking in human cancer. Science, 354:618-622, 2016
2.Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, Shibata T, Naito M, Kim HJ, Miyata K, Kataoka K, Kondo Y. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun, 7:13616, 2016
3.Ojima H, Yamagishi S, Shimada K, Shibata T. Establishment of various biliary tract carcinoma cell lines and xenograft models for appropriate preclinical studies. World J Gastroenterol, 22:9035-9038, 2016
4.Nishimoto Y, Mizutani S, Nakajima T, Hosoda F, Watanabe H, Saito Y, Shibata T, Yachida S, Yamada T. High stability of faecal microbiome composition in guanidine thiocyanate solution at room temperature and robustness during colonoscopy. Gut, 65:1574-1575, 2016
5.Rokutan H, Hosoda F, Hama N, Nakamura H, Totoki Y, Furukawa E, Arakawa E, Ohashi S, Urushidate T, Satoh H, Shimizu H, Igarashi K, Yachida S, Katai H, Taniguchi H, Fukayama M, Shibata T. Comprehensive mutation profiling of mucinous gastric carcinoma. J Pathol, 240:137-148, 2016
6.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, Tanaka H, Chiba K, Ito S, Watatani Y, Kakiuchi N, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Itonaga H, Imaizumi Y, Totoki Y, Munakata W, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Masuda K, Minato N, Kashiwase K, Izutsu K, Takaori-Kondo A, Miyazaki Y, Takahashi S, Shibata T, Kawamoto H, Akatsuka Y, Shimoda K, Takeuchi K, Seya T, Miyano S, Ogawa S. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature, 534:402-406, 2016
7.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet, 48:500-509, 2016
8.Satoh H, Moriguchi T, Saigusa D, Baird L, Yu L, Rokutan H, Igarashi K, Ebina M, Shibata T, Yamamoto M. NRF2 Intensifies Host Defense Systems to Prevent Lung Carcinogenesis, but After Tumor Initiation Accelerates Malignant Cell Growth. Cancer Res, 76:3088-3096, 2016
9.Yachida S, Wood LD, Suzuki M, Takai E, Totoki Y, Kato M, Luchini C, Arai Y, Nakamura H, Hama N, Elzawahry A, Hosoda F, Shirota T, Morimoto N, Hori K, Funazaki J, Tanaka H, Morizane C, Okusaka T, Nara S, Shimada K, Hiraoka N, Taniguchi H, Higuchi R, Oshima M, Okano K, Hirono S, Mizuma M, Arihiro K, Yamamoto M, Unno M, Yamaue H, Weiss MJ, Wolfgang CL, Furukawa T, Nakagama H, Vogelstein B, Kiyono T, Hruban RH, Shibata T. Genomic Sequencing Identifies ELF3 as a Driver of Ampullary Carcinoma. Cancer Cell, 29:229-240, 2016
10.Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y, Iwaya T, Sudo T, Hayashi T, Takai H, Kawasaki Y, Matsukawa T, Eguchi H, Sugimachi K, Tanaka F, Suzuki H, Yamamoto K, Ishii H, Shimizu M, Yamazaki H, Yamazaki M, Tachimori Y, Kajiyama Y, Natsugoe S, Fujita H, Mafune K, Tanaka Y, Kelsell DP, Scott CA, Tsuji S, Yachida S, Shibata T, Sugano S, Doki Y, Akiyama T, Aburatani H, Ogawa S, Miyano S, Mori M, Mimori K. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology, 150:1171-1182, 2016
11.Uchi R, Takahashi Y, Niida A, Shimamura T, Hirata H, Sugimachi K, Sawada G, Iwaya T, Kurashige J, Shinden Y, Iguchi T, Eguchi H, Chiba K, Shiraishi Y, Nagae G, Yoshida K, Nagata Y, Haeno H, Yamamoto H, Ishii H, Doki Y, Iinuma H, Sasaki S, Nagayama S, Yamada K, Yachida S, Kato M, Shibata T, Oki E, Saeki H, Shirabe K, Oda Y, Maehara Y, Komune S, Mori M, Suzuki Y, Yamamoto K, Aburatani H, Ogawa S, Miyano S, Mimori K. Integrated Multiregional Analysis Proposing a New Model of Colorectal Cancer Evolution. PLoS Genet, 12:e1005778, 2016
12.Takai E, Yachida S. Circulating tumor DNA as a liquid biopsy target for detection of pancreatic cancer. World J Gastroenterol, 22:8480-8488, 2016
13.Takai E, Yachida S, Shimizu K, Furuse J, Kubo E, Ohmoto A, Suzuki M, Hruban RH, Okusaka T, Morizane C, Furukawa T. Germline mutations in Japanese familial pancreatic cancer patients. Oncotarget, 7:74227-74235, 2016
14.Nagata Y, Kontani K, Enami T, Kataoka K, Ishii R, Totoki Y, Kataoka TR, Hirata M, Aoki K, Nakano K, Kitanaka A, Sakata-Yanagimoto M, Egami S, Shiraishi Y, Chiba K, Tanaka H, Shiozawa Y, Yoshizato T, Suzuki H, Kon A, Yoshida K, Sato Y, Sato-Otsubo A, Sanada M, Munakata W, Nakamura H, Hama N, Miyano S, Nureki O, Shibata T, Haga H, Shimoda K, Katada T, Chiba S, Watanabe T, Ogawa S. Variegated RHOA mutations in adult T-cell leukemia/lymphoma. Blood, 127:596-604, 2016
15.Luchini C, Veronese N, Yachida S, Cheng L, Nottegar A, Stubbs B, Solmi M, Capelli P, Pea A, Barbareschi M, Fassan M, Wood LD, Scarpa A. Different prognostic roles of tumor suppressor gene BAP1 in cancer: A systematic review with meta-analysis. Genes Chromosomes Cancer, 55:741-749, 2016
16.Ohmoto A, Yachida S, Kubo E, Takai E, Suzuki M, Shimada K, Okusaka T, Morizane C. Clinicopathologic Features and Germline Sequence Variants in Young Patients (≤40 Years Old) With Pancreatic Ductal Adenocarcinoma. Pancreas, 45:1056-1061, 2016.
17.Matsuda Y, Ishiwata T, Yachida S, Suzuki A, Hamashima Y, Hamayasu H, Yoshimura H, Honma N, Aida J, Takubo K, Arai T. Clinicopathological Features of 15 Occult and 178 Clinical Pancreatic Ductal Adenocarcinomas in 8339 Autopsied Elderly Patients. Pancreas, 45:234-240, 2016
18.Ichimura K, Fukushima S, Totoki Y, Matsushita Y, Otsuka A, Tomiyama A, Niwa T, Takami H, Nakamura T, Suzuki T, Fukuoka K, Yanagisawa T, Mishima K, Nakazato Y, Hosoda F, Narita Y, Shibui S, Yoshida A, Mukasa A, Saito N, Kumabe T, Kanamori M, Tominaga T, Kobayashi K, Shimizu S, Nagane M, Iuchi T, Mizoguchi M, Yoshimoto K, Tamura K, Maehara T, Sugiyama K, Nakada M, Sakai K, Kanemura Y, Nonaka M, Asai A, Yokogami K, Takeshima H, Kawahara N, Takayama T, Yao M, Kato M, Nakamura H, Hama N, Sakai R, Ushijima T, Matsutani M, Shibata T, Nishikawa R. Recurrent neomorphic mutations of MTOR in central nervous system and testicular germ cell tumors may be targeted for therapy. Acta Neuropathol, 131:889-901, 2016
19.Kubota D, Kosaka N, Fujiwara T, Yoshida A, Arai Y, Qiao Z, Takeshita F, Ochiya T, Kawai A, Kondo T. miR-125b and miR-100 Are Predictive Biomarkers of Response to Induction Chemotherapy in Osteosarcoma. Sarcoma, 2016:1390571, 2016
20.Mimaki S, Totsuka Y, Suzuki Y, Nakai C, Goto M, Kojima M, Arakawa H, Takemura S, Tanaka S, Marubashi S, Kinoshita M, Matsuda T, Shibata T, Nakagama H, Ochiai A, Kubo S, Nakamori S, Esumi H, Tsuchihara K. Hypermutation and unique mutational signatures of occupational cholangiocarcinoma in printing workers exposed to haloalkanes. Carcinogenesis, 37:817-826, 2016