HOME > Publication & Reports > Annual Report 2016 > Research Institute
Division of Genome Biology
Takashi Kohno, Naoto Tsuchiya, Hideaki Ogiwara, Kouya Shiraishi, Takashi Nakaoku, Kuniko Sunami, Ken Ishida, Mariko Sasaki, Yoko Shimada, Kazuaki Takahashi, Yuko Fujiwara, Takayuki Honda, Yuko Nishiyama, Kazushi Yoshida, Yoshie Iga, Ayako Otsuka, Maiko Matsuda, Keisuke Sugiyama
Introduction
The clarification of somatic mutation signatures in the cancer genome and inter-individual genetic polymorphisms in the human genome is crucial key to improve cancer medicine. The Division of Genome Biology aims to find "seeds" that are applicable for the development of novel strategies for treatment, diagnosis, and prevention of cancer through identifying and understanding the biological relationship of their seeds with cancer pathogenesis caused by somatic mutations and/or genetic polymorphisms of patients. In order to attain our goal, we are working together with the National Cancer Center (NCC) staff from the Hospital, the Exploratory Oncology Research & Clinical Trial Center (EPOC), and the Center for Public Health Sciences to fight lung cancer, the most common cause of cancer-related death worldwide.
Our team and what we do
1)Genome research team; finding unique seeds applicable for cancer medicine.
2)miRNA team; understanding molecular networks in cancer cells
Research activities
1.Genes for personalized cancer medicine
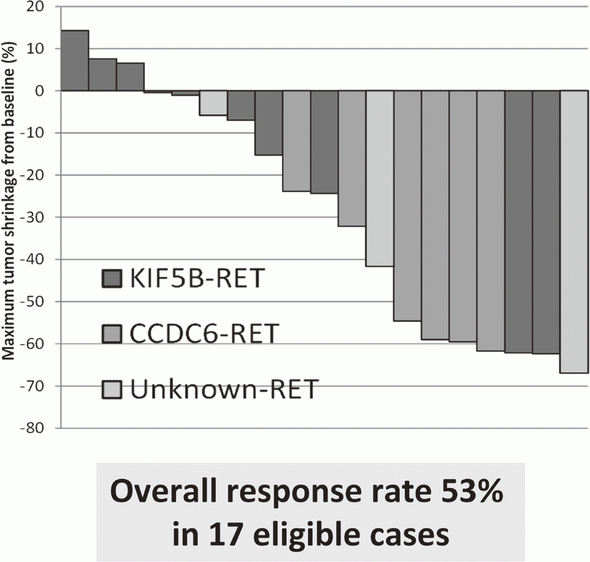
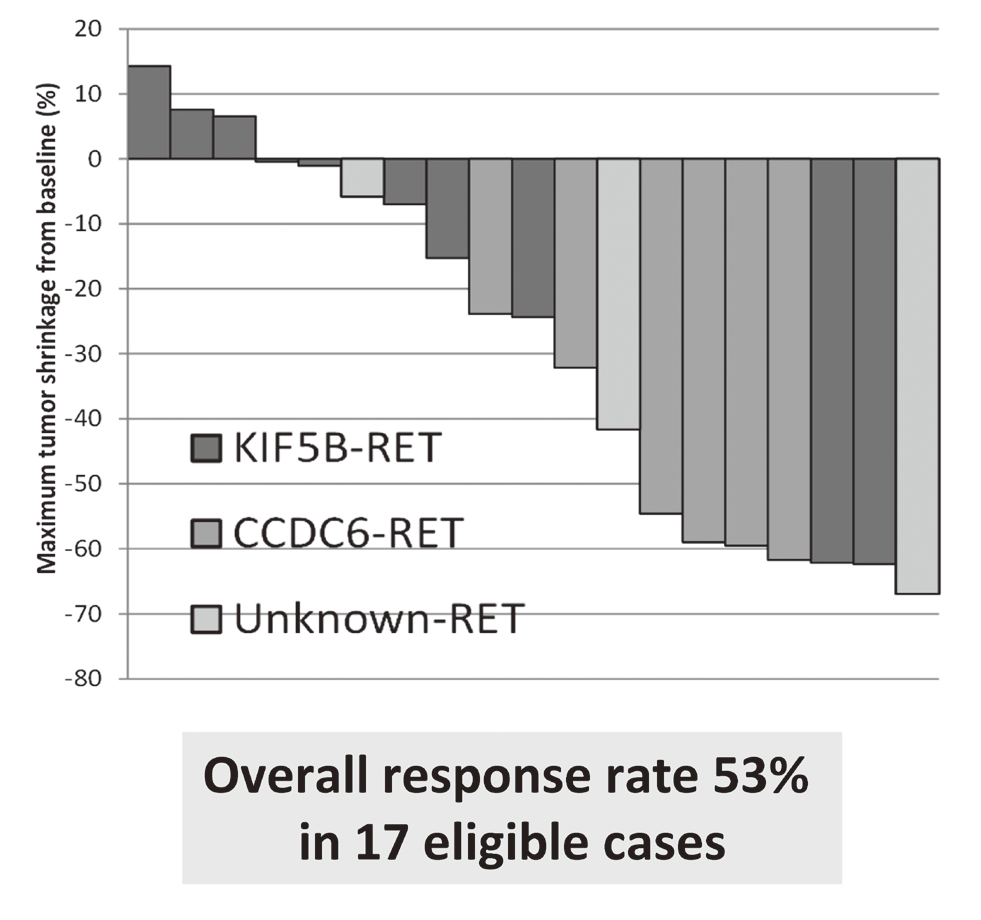
LURET, an open-label, multicenter phase II trial of Vandetanib in patients with RET-rearranged advanced non-small-cell lung cancer revealed that RET fusion, a new driver oncogene that we found, is a promising therapeutic target (Figure 1). Synthetic lethality therapy for CREBBP-deficient small cell lung cancer was proposed; EP300 histone acetyltransferase is a vulnerability of such cancer, and its functional suppression causes depletion of MYC oncogene products. Patient-derived ovarian cancer cells were cultured and used for synthetic lethal screening to identify therapeutic targets for cancers deficient for SWI/SNF chromatin remodelers. NRG1 fusion gene, that we discovered to be a new driver oncogene, was shown to increase cancer stem-like property through enhancing the signaling circuit of the NFKB-IGF2 pathway.
Risk factors for lung adeno-carcinogenesis were searched for by genome-wide studies. Genome-wide association study (GWAS) of LADC with EGFR mutation was performed as a multi-center collaborative study with Japanese population consisting of 3,173 cases and 15,158 controls. Six loci, including HLA-DPB1, were discovered as risk loci for the disease. International GWAS uncovered multiple loci associated with a lung cancer risk of revealed mutation burden caused by multiple factors, including smoking. To understand mutational and/or genetic features of cancers developed in adolescents and young adults (AYA; 15-39 years old), genome-wide analyses were launched for ovarian, breast, uterine, and lung tumors.
2.Identification of molecular target networks based on the property of miRNA
Tumor-suppressive miR-101, which was identified by functional screening, is found to be a crucial factor in p53-dependent nucleolus stress pathway resulted in cells undergoing apoptosis. miR-101 was down-regulated in various cancers, including lung and colon cancers. Importantly, low expression of miR-101 was significantly associated with poor prognosis exclusively in p53-wild type LADC, strongly suggesting that miR-101 participates in intrinsic tumor suppression in cooperation with p53. Reactivation of this circuit is suggested to be a promising strategy for cancer treatment. The miRNA team (Dr. Tsuchiya's team) will be independent in April 2017 as the "Laboratory of Molecular Carcinogenesis".
Clinical trials
The results of a phase II clinical trial (LURET), which investigates the therapeutic effect of a RET-tyrosine kinase inhibitor, and vandetanib were reported. The overall response rate was 53% (Figure 1).
Education
Supervising researches and presentation skills for graduate school students and young researchers.
Future prospects
The goal of our division is the establishment of novel strategies for personalized cancer medicine through the finding of unique "seeds". Genome analyses, including those of AYA-cancers, will contribute to precision prevention and therapy of cancers. Understanding biological roles of novel molecular targets, which have been identified by synthetic lethal screen and comprehensive genome analyses, will provide unique and novel concepts improving cancer therapy.
List of papers published in 2016
Journal
1.Saito M, Shiraishi K, Kunitoh H, Takenoshita S, Yokota J, Kohno T. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer Sci, 107:713-720, 2016
2.Kato F, Fiorentino FP, Alibes A, Perucho M, Sanchez-Cespedes M, Kohno T, Yokota J. MYCL is a target of a BET bromodomain inhibitor, JQ1, on growth suppression efficacy in small cell lung cancer cells. Oncotarget, 7:77378-77388, 2016
3.Anadon C, Guil S, Simo-Riudalbas L, Moutinho C, Setien F, Martinez-Cardus A, Moran S, Villanueva A, Calaf M, Vidal A, Lazo PA, Zondervan I, Savola S, Kohno T, Yokota J, Ribas de Pouplana L, Esteller M. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene, 35:4422, 2016
4.Fiorentino FP, Tokgun E, Sole-Sanchez S, Giampaolo S, Tokgun O, Jauset T, Kohno T, Perucho M, Soucek L, Yokota J. Growth suppression by MYC inhibition in small cell lung cancer cells with TP53 and RB1 inactivation. Oncotarget, 7:31014-31028, 2016
5.Shi J, Hua X, Zhu B, Ravichandran S, Wang M, Nguyen C, Brodie SA, Palleschi A, Alloisio M, Pariscenti G, Jones K, Zhou W, Bouk AJ, Boland J, Hicks B, Risch A, Bennett H, Luke BT, Song L, Duan J, Liu P, Kohno T, Chen Q, Meerzaman D, Marconett C, Laird-Offringa I, Mills I, Caporaso NE, Gail MH, Pesatori AC, Consonni D, Bertazzi PA, Chanock SJ, Landi MT. Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study. PLoS Med, 13:e1002162, 2016
6.Seki Y, Fujiwara Y, Kohno T, Takai E, Sunami K, Goto Y, Horinouchi H, Kanda S, Nokihara H, Watanabe S, Ichikawa H, Yamamoto N, Kuwano K, Ohe Y. Picoliter-Droplet Digital Polymerase Chain Reaction-Based Analysis of Cell-Free Plasma DNA to Assess EGFR Mutations in Lung Adenocarcinoma That Confer Resistance to Tyrosine-Kinase Inhibitors. Oncologist, 21:156-164, 2016
7.Shiraishi K, Okada Y, Takahashi A, Kamatani Y, Momozawa Y, Ashikawa K, Kunitoh H, Matsumoto S, Takano A, Shimizu K, Goto A, Tsuta K, Watanabe S, Ohe Y, Watanabe Y, Goto Y, Nokihara H, Furuta K, Yoshida A, Goto K, Hishida T, Tsuboi M, Tsuchihara K, Miyagi Y, Nakayama H, Yokose T, Tanaka K, Nagashima T, Ohtaki Y, Maeda D, Imai K, Minamiya Y, Sakamoto H, Saito A, Shimada Y, Sunami K, Saito M, Inazawa J, Nakamura Y, Yoshida T, Yokota J, Matsuda F, Matsuo K, Daigo Y, Kubo M, Kohno T. Association of variations in HLA class II and other loci with susceptibility to EGFR-mutated lung adenocarcinoma. Nat Commun, 7:12451, 2016
8.Kamata T, Sunami K, Yoshida A, Shiraishi K, Furuta K, Shimada Y, Katai H, Watanabe S, Asamura H, Kohno T, Tsuta K. Frequent BRAF or EGFR mutations in ciliated muconodular papillary tumors of the lung. J Thorac Oncol, 11:261-265, 2016
9.Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, Kohno T. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell Death due to abrogation of MYC expression. Cancer Discov, 6:430-445, 2016
10.Murayama T, Nakaoku T, Enari M, Nishimura T, Tominaga K, Nakata A, Tojo A, Sugano S, Kohno T, Gotoh N. Oncogenic fusion gene CD74-NRG1 confers cancer stem cell-like properties in lung cancer through a IGF2 autocrine/paracrine circuit. Cancer Res, 76:974-983, 2016
11.Sunami K, Furuta K, Tsuta K, Sasada S, Izumo T, Nakaoku T, Shimada Y, Saito M, Nokihara H, Watanabe S, Ohe Y, Kohno T. Multiplex Diagnosis of Oncogenic Fusion and MET Exon Skipping by Molecular Counting Using Formalin-Fixed Paraffin Embedded Lung Adenocarcinoma Tissues. J Thorac Oncol, 11:203-212, 2016
12.Asao T, Fujiwara Y, Sunami K, Kitahara S, Goto Y, Kanda S, Horinouchi H, Nokihara H, Yamamoto N, Ichikawa H, Kohno T, Tsuta K, Watanabe S, Takahashi K, Ohe Y. Medical treatment involving investigational drugs and genetic profile of thymic carcinoma. Lung Cancer, 93:77-81, 2016
13.Wang Z, Seow WJ, Shiraishi K, Hsiung CA, Matsuo K, Liu J, Chen K, Yamji T, Yang Y, Chang IS, Wu C, Hong Y-C, Burdett L, Wyatt K, Chung CC, Li SA, Yeager M, Hutchinson A, Hu W, Caporaso N, Landi MT, Chatterjee N, Song M, Fraumeni JF, Jr., Kohno T, Yokota J, Kunitoh H, Ashikawa K, Momozawa Y, Daigo Y, Mitsudomi T, Yatabe Y, Hida T, Hu Z, Dai J, Ma H, Jin G, Song B, Wang Z, Cheng S, Yin Z, Li X, Ren Y, Guan P, Chang J, Tan W, Chen C-J, Chang G-C, Tsai Y-H, Su W-C, Chen K-Y, Huang M-S, Chen Y-M, Zheng H, Li H, Cui P, Guo H, Xu P, Liu L, Iwasaki M, Shimazu T, Tsugane S, Zhu J, Jiang G, Fei K, Park JY, Kim YH, Sung JS, Park KH, Kim YT, Jung YJ, Kang CH, Park IK, Kim HN, Jeon H-S, Choi JE, Choi YY, Kim JH, Oh I-J, Kim Y-C, Sung SW, Kim JS, Yoon H-I, Kweon S-S, Shin M-H, Seow A, Chen Y, Lim W-Y, Liu J, Wong MP, Lee VHF, Bassig BA, Tucker M, Berndt SI, Chow W-H, Ji B-T, Wang J, Xu J, Sihoe ADL, Ho JCM, Chan JKC, Wang J-C, Lu D, Zhao X, Zhao Z, Wu J, Chen H, Jin L, Wei F, Wu G, An S-J, Zhang X-C, Su J, Wu Y-L, Gao Y-T, Xiang Y-B, He X, Li J, Zheng W, Shu X-O, Cai Q, Klein R, Pao W, Lawrence C, Hosgood HD, 3rd, Hsiao C-F, Chien L-H, Chen Y-H, Chen C-H, Wang W-C, Chen C-Y, Wang C-L, Yu C-J, Chen H-L, Su Y-C, Tsai F-Y, Chen Y-S, Li Y-J, Yang T-Y, Lin C-C, Yang P-C, Wu T, Lin D, Zhou B, Yu J, Shen H, Kubo M, Chanock SJ, Rothman N, Lan Q. Meta-analysis of genome-wide association studies identifies multiple lung cancer susceptibility loci in never-smoking Asian women. Hum Mol Genet, 25:620-629, 2016
14.Tanabe Y, Ichikawa H, Kohno T, Yoshida H, Kubo T, Kato M, Iwasa S, Ochiai A, Yamamoto N, Fujiwara Y, Tamura K. Comprehensive screening of target molecules by next-generation sequencing in patients with malignant solid tumors: guiding entry into phase I clinical trials. Mol Cancer, 15:73, 2016