HOME > Publication & Reports > Annual Report 2016 > Research Institute
Division of Molecular Modification and Cancer Biology
Ryuji Hamamoto, Syuzo Kaneko, Noriko Ikawa, Kyoko Fujioka, Hidenori Machino, Shinya Oki, Machiko Kojima, Takeaki Dohda, Reiko Masuhara
Introduction
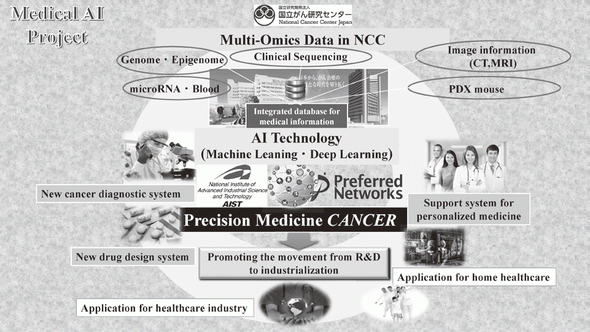
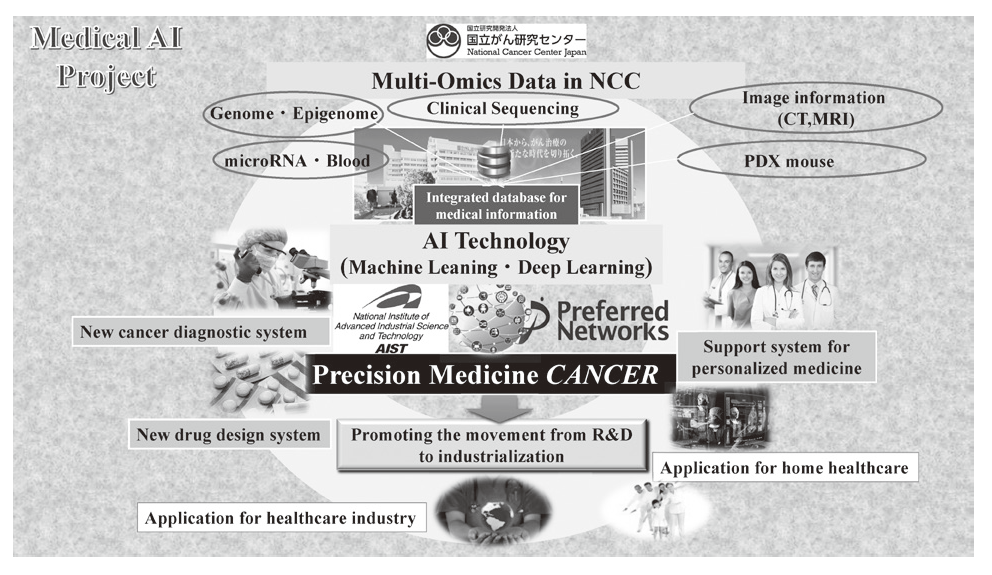
In 2004, we reported the functions of SMYD3, a histone methyltransferase, in human tumorigenesis in Nature Cell Biology, and this was the first report to describe how dysregulation of histone methylation contributes to human tumorigenesis at the molecular level. This article was also introduced in Nature Reviews Cancer and Nature Reviews Molecular Cell Biology, and the press release was issued in Nature Cell Biology because the report showed a novel type of target for anti-cancer therapy. Since then, we have already reported a number of enzymes relevant to protein methylation, which are involved in human cancer, and contributes to the progress of novel field of cancer research. Dr. Hamamoto, the chief of the Division of Molecular Modification and Cancer Biology, wrote an article describing our previous achievements in Nature Reviews Cancer (Hamamoto et al., Nature Reviews Cancer, 15, 110 - 124 [2015]) as the first and corresponding author. We also started a new project relevant to cancer medical AI in 2016
Research activities
1)We identified three novel substrates of non-histone methylation as shown below.
a.AKT1 methylation by histone methyltransferase SMYD3 to regulate the PI3K-AKT1 pathway
b.Automethylation of the histone methyltransferase SUV39H2 to regulate its enzyme activity
c.EGFR methylation by histone methyltransferase WHSC1L1 to regulate the downstream pathway
2)We demonstrated that expression levels of the histone methyltransferase EZH2 and the immune checkpoint blockade agent PD-L1 were statistically correlated in lung cancer patients (adenocarcinoma, squamous carcinoma, small cell carcinoma). In addition, lung cancer patients with EZH2+/PD-L1+ showed poorer prognosis than those with EZH2+/PD-L1- (both recurrence-free survival and overall survival).
3)We are developing the next generation ChIP-Seq method using machine learning to analyze the status of histone modifications and the DNA binding condition of transcription factors, which can also analyze FFPE samples and low cell number samples.
Education
Three graduate school students from the Graduate School of Medicine, the University of Tokyo were trained in our division.
Future prospects
1)Establishment of epiproteomics field
a.The next generation ChIP-Seq method using advanced technologies such as machine learning and deep learning
b.Combination of mass-spectrometry and CRISPR system to analyze the status of histone modifications without the bias of antibody quality
2)Development of an integrated medical system for cancer treatment and diagnosis using AI technology
3)Development of a novel molecular targeted therapy targeting histone methyltransferase and demethylase
List of papers published in 2016
Journal
1.Hamamoto R, Nakamura Y. Dysregulation of protein methyltransferases in human cancer: An emerging target class for anticancer therapy. Cancer Sci, 107:377-384, 2016
2.Piao L, Nakakido M, Suzuki T, Dohmae N, Nakamura Y, Hamamoto R. Automethylation of SUV39H2, an oncogenic histone lysine methyltransferase, regulates its binding affinity to substrate proteins. Oncotarget, 7:22846-22856, 2016
3.Saloura V, Vougiouklakis T, Zewde M, Kiyotani K, Park J-H, Gao G, Karrison T, Lingen M, Nakamura Y, Hamamoto R. WHSC1L1 drives cell cycle progression through transcriptional regulation of CDC6 and CDK2 in squamous cell carcinoma of the head and neck. Oncotarget, 7:42527-42538, 2016
4.Kunizaki M, Sawai T, Takeshita H, Tominaga T, Hidaka S, To K, Miyazaki T, Hamamoto R, Nanashima A, Nagayasu T. Clinical Value of Serum p53 Antibody in the Diagnosis and Prognosis of Colorectal Cancer. Anticancer Res, 36:4171-4175, 2016
5.Toyokawa G, Taguchi K, Edagawa M, Shimamatsu S, Toyozawa R, Nosaki K, Hirai F, Yamaguchi M, Shimokawa M, Seto T, Takenoyama M, Hamamoto R, Sugio K, Ichinose Y. The Prognostic Impact of Jumonji Domain-containing 2B in Patients with Resected Lung Adenocarcinoma. Anticancer Res, 36:4841-4846, 2016
6.Yoshioka Y, Suzuki T, Matsuo Y, Nakakido M, Tsurita G, Simone C, Watanabe T, Dohmae N, Nakamura Y, Hamamoto R. SMYD3-mediated lysine methylation in the PH domain is critical for activation of AKT1. Oncotarget, 7:75023-75037, 2016
7.Deng Y, Wang H, Hamamoto R, Duan S, Pirooznia M, Bai Y. Functional Genomics, Genetics, and Bioinformatics 2016. Biomed Res Int, 2016:2625831, 2016