HOME > Publication & Reports > Annual Report 2016 > Research Institute
Department of Innovative Seeds Evaluation
Tadashi Kondo, Tsutomu Ohta, Rieko Oyama, Yoko Takai, Fusako Kito, Marimu Sakumoto, Shoko Tanahashi, Naoya Hirai
Introduction
We focus on two projects; the establishment of a patient-derived cancer model of rare cancers, and the application of developed cancer models for the preclinical study. The patient-derived cancer model is an essentially important tool to understand the disease backgrounds, and develop novel therapeutic strategies. Many anti-cancer drugs and biomarkers have been developed using the patient-derived cancer models. However, the patient-derived cancer models are available only in a limited portion of cancers. For example, they are hardly available for rare cancers. With this notion, we launched the project to establish the patient-derived cancer models. One of the most important applications of the patient-derived cancer models is the examination of efficacy of anti-cancer drugs as a pre-clinical study. With rare cancers, the pre-clinical study has been hampered because of the lack of patient-derived cancer models. Thus, we created the evaluation system for the efficacy of anti-cancer drugs using our original patient-derived cancer models.
Research activities
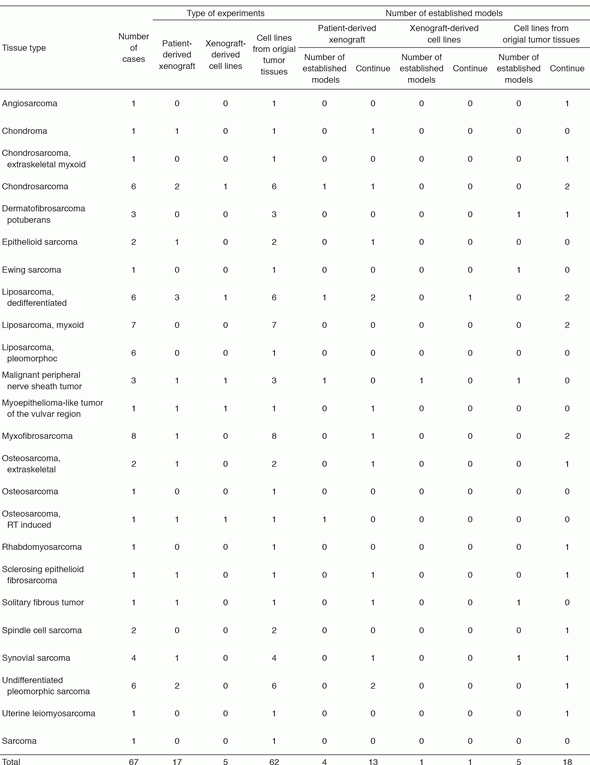
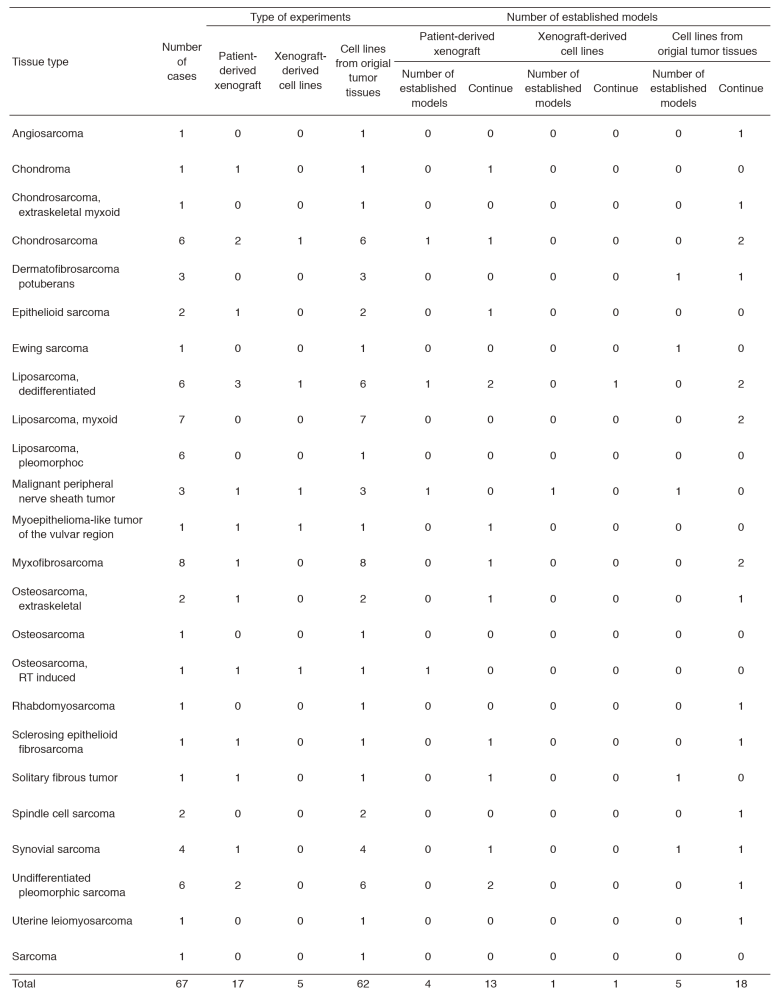
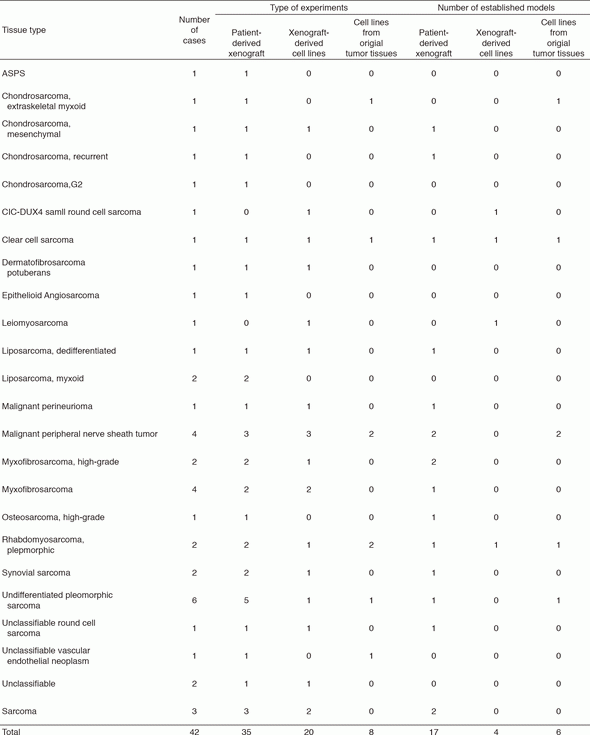
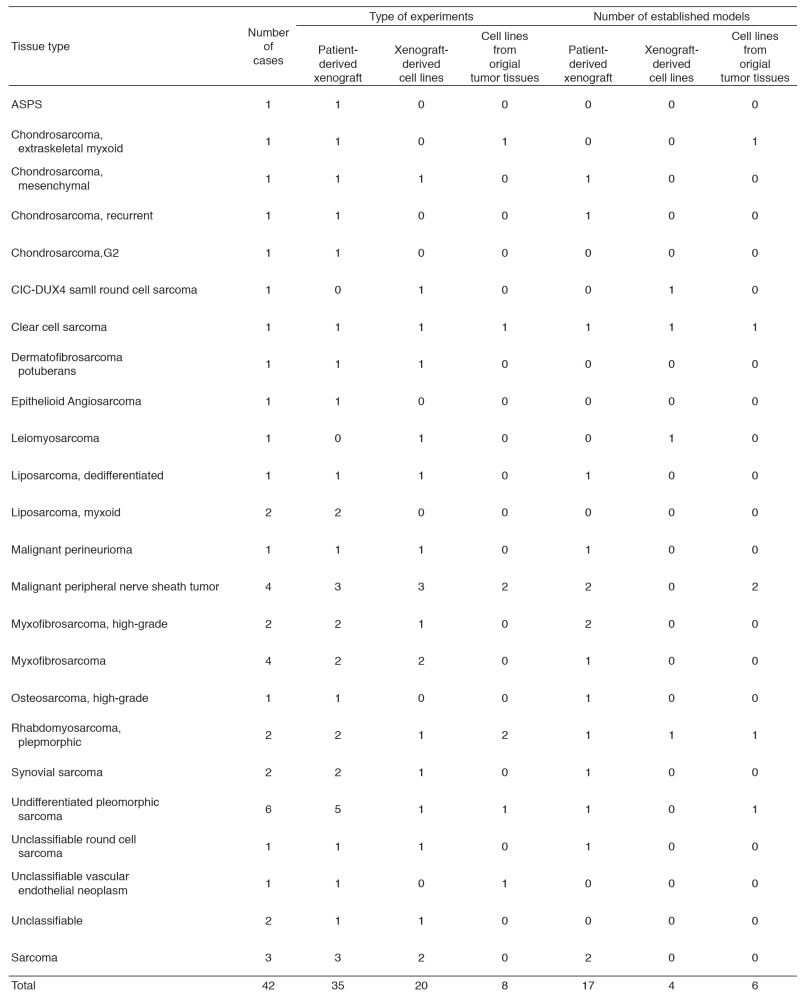
The surgically dissected tumor tissues obtained in the National Cancer Center Hospital were used to develop the patient-derived cancer models. The tumor tissues are quite diverse in terms of substances in the tissues and heterogeneity of cell populations depending on the original tissue samples, and the methods for individual histology are required to establish the cancer models. We established the patient-derived sarcoma models (Tables 1 and 2). The molecular characterization is also important to use the cancer models in the research. Because the molecular backgrounds are altered during the process of cancer model establishment, it is quite important to know how the developed cancer models retain the original molecular backgrounds. For this sake, we employ the multi-omics approach. The DNA, RNA, and proteins are comprehensively and intensively examined, comparing the original tissue samples and the established models. Our cancer models are included in the collaborative study with pharmaceutical companies. The research activity of our department is linked to that of the Division of Rare Cancer Research. The ideas and the fundamental research tools are shared between two laboratories for novel innovative seeds development. In collaboration with the Division of Rare Cancer Research, we screened more than 150 anti-cancer drugs using our original patient-derived cancer models.
Future prospests
Our research activities will benefit patients with rare cancers by contributing to the development of novel cancer drugs. We will establish more collaborative studies with pharmaceutical companies as well as academic research groups.
List of papers published in 2016
Journal
1.Ezawa I, Sawai Y, Kawase T, Okabe A, Tsutsumi S, Ichikawa H, Kobayashi Y, Tashiro F, Namiki H, Kondo T, Semba K, Aburatani H, Taya Y, Nakagama H, Ohki R. Novel p53 target gene FUCA1 encodes a fucosidase and regulates growth and survival of cancer cells. Cancer Sci, 107:734-745, 2016
2.Asano Y, Kawase T, Okabe A, Tsutsumi S, Ichikawa H, Tatebe S, Kitabayashi I, Tashiro F, Namiki H, Kondo T, Semba K, Aburatani H, Taya Y, Nakagama H, Ohki R. IER5 generates a novel hypo-phosphorylated active form of HSF1 and contributes to tumorigenesis. Sci Rep, 6:19174, 2016
3.Kondo T. Proteogenomics for the Study of Gastrointestinal Stromal Tumors. Adv Exp Med Biol, 926:139-151, 2016
4.Morofuji N, Ojima H, Hiraoka N, Okusaka T, Esaki M, Nara S, Shimada K, Kishi Y, Kondo T. Antibody-based proteomics to identify an apoptosis signature for early recurrence of hepatocellular carcinoma. Clin Proteomics, 13:28, 2016
5.Takai Y, Matsuo A, Qiao Z, Kondo T. Antitumor activities of BIBF 1120, BI 860585, and BI 836845 in preclinical models of sarcoma. Integr Mol Med, 3:755-760, 2016
6.Yamasaki H, Miyamoto M, Yamamoto Y, Kondo T, Watanabe T, Ohta T. Synovial sarcoma cell lines showed reduced DNA repair activity and sensitivity to a PARP inhibitor. Genes Cells, 21:852-860, 2016
7.Pan X, Yoshida A, Kawai A, Kondo T. Current status of publicly available sarcoma cell lines for use in proteomic studies. Expert Rev Proteomics, 13:227-240, 2016
8.Kubota D, Kosaka N, Fujiwara T, Yoshida A, Arai Y, Qiao Z, Takeshita F, Ochiya T, Kawai A, Kondo T. miR-125b and miR-100 Are Predictive Biomarkers of Response to Induction Chemotherapy in Osteosarcoma. Sarcoma, 2016:1390571, 2016