Annual Report 2017
Division of Cancer Immunology
Hiroyoshi Nishikawa, Yosuke Togashi, Chika Sakai, Yasuko Tada, Eri Sugiyama, Shota Fukuoka, Tomomi Morioka, Yoko Ohira, Konomi Onagawa, Tomoka Takaku, Yoshiko Takeuchi, Megumi Takemura, Miyuki Nakai, Chie Haijima, Yuki Fujioka, Megumi Hoshino, Kumiko Yoshida, Sayuri Yoshimatsu
Introduction
Recent success of cancer immunotherapy, particularly immune checkpoint inhibitors, makes it a key strategy for cancer treatment and a lot of clinical trials are under investigation for developing new reagents. Among patients receiving cancer immunotherapy, while some experience long-lasting clinical benefits, more than half of the patients fail to exhibit clinical benefit. Moreover, patients sometimes experience immune-related adverse effects such as thyroiditis and pneumonitis even without clinical benefits. It is therefore urgently required to identify predictive biomarkers, and to develop more effective therapies and optimal immunotherapies for individual patients.
Our team and what we do
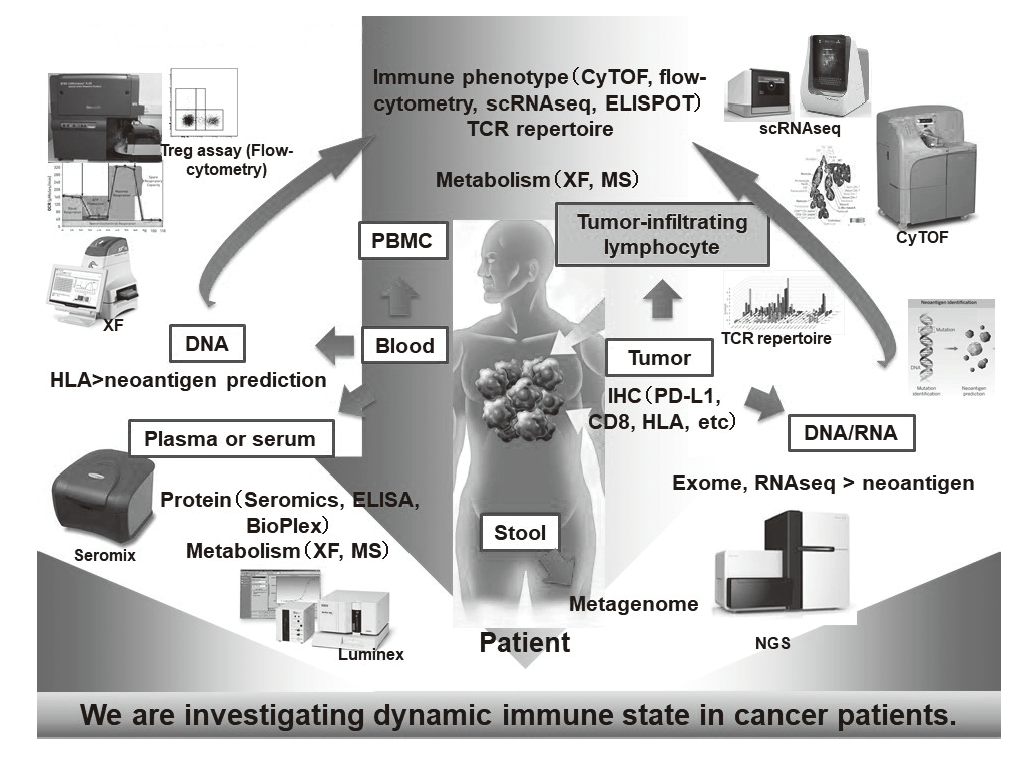
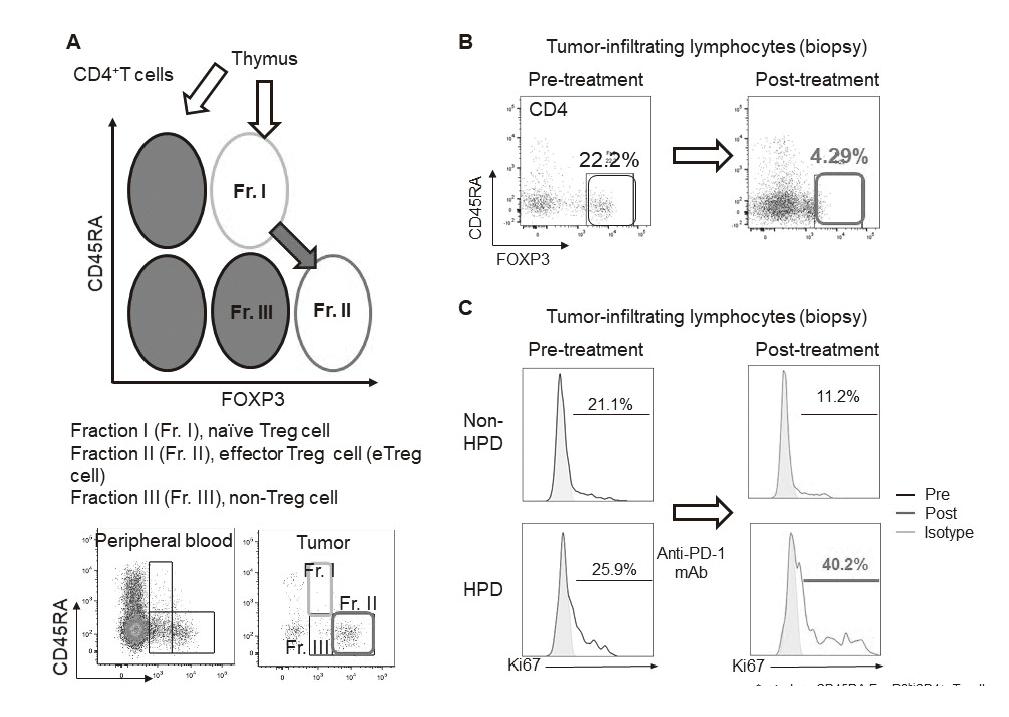
In our laboratory, viable immune cells both in tumors and in the periphery are being investigated using several techniques such as multi-color flow-cytometry, CyTOF, and single cell RNA sequencing in collaboration with clinical department (Figure 1). A real-time immune monitoring system in cancer patients during their cancer treatment has been established, by which we have had collaborations with pharmaceutical companies in their clinical trials. Especially, regulatory T cells (Treg cells), which have a strong immune suppressive function and are known to be a critical component to inhibit anti-tumor immunity, are analyzed with our original assay (effector Treg cells; eTreg cells) (Figure 2A). Furthermore, with the findings from clinical sample analyses, we also reverse-translate into basic research to elucidate the essential mechanisms of immune responses to cancer.
Figure 1. Clinical sample analysis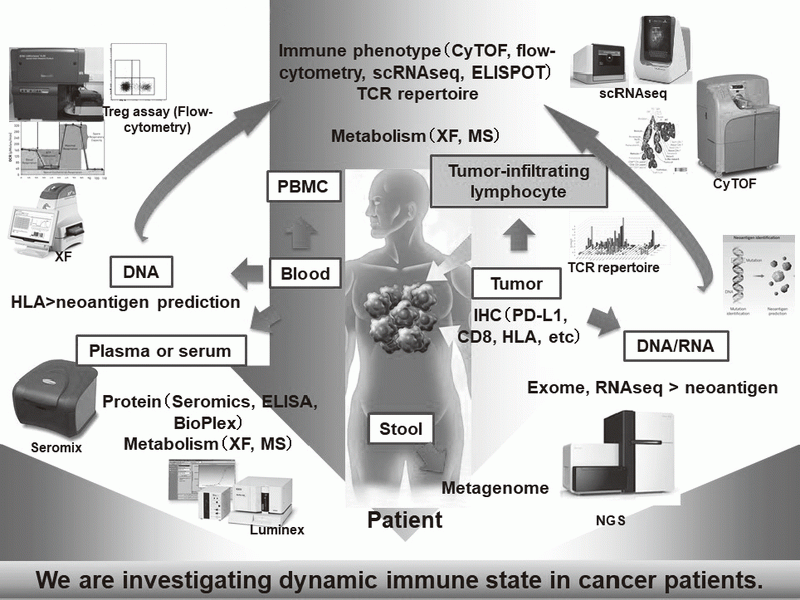
Research activities
Various types of cancers have been classified into subgroups based on genomic alterations; meanwhile, a classification by genomic analyses has shown limitations to elucidate detailed immune status in tumor microenvironment due to the complexity of the immune system. When the data from our assays of immune status in tumor microenvironment were integrated with genomic analyses in non-small cell lung cancer (NSCLC), significant differences between EGFR-mutated and wild-type NSCLC were identified (manuscript in preparation). In addition, we have found a specific driver mutation in gastric cancer which influenced immune status in tumor microenvironment (manuscript in preparation).
Using our real-time immune monitoring system, we also addressed kinetic changes of immune status in tumor microenvironment after cancer therapies including not only immune checkpoint inhibitors, but also cytotoxic reagents. We found a reduction of eTreg cells by a first-in-human clinical reagent using our real-time immune monitoring system, proposing a novel effect of this drug (Figure 2B). Furthermore, a specific immune state change in hyperprogressive disease (HPD) (Figure 2C) and a possible predictive biomarker have been identified in patients treated with immune checkpoint inhibitors. We have also worked on GAPFREE2 project in which tumor-infiltrating lymphocytes at both pre- and post-PD-1 blockade are analyzed using scRNAseq.
Clinical trials
We have collaborated with many pharmaceutical companies in their clinical trials (chemoradiotherapy/nivolumab neo-adjuvant, BBI608/pembrolizumab, OBP301/pembrolizumab, ipilimumab/nivolumab, regorafenib/nivolumab, atezolizumab, and etc.) to elucidate predictive biomarkers and immune status in tumor microenvironment using our real-time immune monitoring system.
Education
Seven graduate school students are trained in our division, and two new students will join in the next year. After finishing their theses, they continue to study cancer immunology abroad (Icahn SOM at Mount Sinai, Univ. of Michigan and Washington Univ.). Young residents are also trained in our division to become physician scientists.
Future prospects
We integrate our immunological assay data in tumor microenvironment with genomic assay data in many cancer types, resulting in establishment of "cancer immune genome atlas." Mechanistic studies are also addressed based on these clinical findings. Our real-time immune monitoring system in cancer patients pre-, during and post-cancer treatment is supposed to be attractive and appreciated by a lot of pharmaceutical and medical device companies for effective drug development. Many collaborative studies with such companies associated with clinical trials are therefore ongoing and being planned, which will become a valuable framework for translational and reverse-translational research.
List of papers published in January 2017 - March 2018
Journal
1. Enokida T, Nishikawa H. Regulatory T cells, as a target in anticancer immunotherapy. Immunotherapy, 9:623-627, 2017
2. Kasahara H, Kondo T, Nakatsukasa H, Chikuma S, Ito M, Ando M, Kurebayashi Y, Sekiya T, Yamada T, Okamoto S, Yoshimura A. Generation of allo-antigen-specific induced Treg stabilized by vitamin C treatment and its application for prevention of acute graft versus host disease model. Int Immunol, 29:457-469, 2017
3. Togashi Y, Nishikawa H. Suppression from beyond the grave. Nat Immunol, 18:1285-1286, 2017
4. Doki N, Suyama M, Sasajima S, Ota J, Igarashi A, Mimura I, Morita H, Fujioka Y, Sugiyama D, Nishikawa H, Shimazu Y, Suda W, Takeshita K, Atarashi K, Hattori M, Sato E, Watakabe-Inamoto K, Yoshioka K, Najima Y, Kobayashi T, Kakihana K, Takahashi N, Sakamaki H, Honda K, Ohashi K. Clinical impact of pre-transplant gut microbial diversity on outcomes of allogeneic hematopoietic stem cell transplantation. Ann Hematol, 96:1517-1523, 2017
5. Haratani K, Hayashi H, Tanaka T, Kaneda H, Togashi Y, Sakai K, Hayashi K, Tomida S, Chiba Y, Yonesaka K, Nonagase Y, Takahama T, Tanizaki J, Tanaka K, Yoshida T, Tanimura K, Takeda M, Yoshioka H, Ishida T, Mitsudomi T, Nishio K, Nakagawa K. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation-positive non-small-cell lung cancer based on T790M status after disease progression during EGFR-TKI treatment. Ann Oncol, 28:1532-1539, 2017
6. Chiba M, Togashi Y, Bannno E, Kobayashi Y, Nakamura Y, Hayashi H, Terashima M, De Velasco MA, Sakai K, Fujita Y, Mitsudomi T, Nishio K. Efficacy of irreversible EGFR-TKIs for the uncommon secondary resistant EGFR mutations L747S, D761Y, and T854A. BMC Cancer, 17:281, 2017
7. Nishiwaki S, Sugiura I, Miyata Y, Saito S, Sawa M, Nishida T, Miyamura K, Kuwatsuka Y, Kohno A, Yuge M, Kasai M, Iida H, Kurahashi S, Osaki M, Goto T, Terakura S, Murata M, Nishikawa H, Kiyoi H. Efficacy and safety of autologous peripheral blood stem cell transplantation for Philadelphia chromosome-positive acute lymphoblastic leukemia: A study protocol for a multicenter exploratory prospective study (Auto-Ph17 study). Medicine (Baltimore), 96:e9568, 2017
8. Banno E, Togashi Y, de Velasco MA, Mizukami T, Nakamura Y, Terashima M, Sakai K, Fujita Y, Kamata K, Kitano M, Kudo M, Nishio K. Clinical significance of Akt2 in advanced pancreatic cancer treated with erlotinib. Int J Oncol, 50:2049-2058, 2017
9. Togashi Y, Nishikawa H. Regulatory T Cells: Molecular and Cellular Basis for Immunoregulation. Current topics in microbiology and immunology, 410:43186, 2017
10. Nagase H, Takeoka T, Urakawa S, Morimoto-Okazawa A, Kawashima A, Iwahori K, Takiguchi S, Nishikawa H, Sato E, Sakaguchi S, Mori M, Doki Y, Wada H. ICOS+ Foxp3+ TILs in gastric cancer are prognostic markers and effector regulatory T cells associated with Helicobacter pylori. Int J Cancer, 140:686-695, 2017
11. Hamano Y, Kida H, Ihara S, Murakami A, Yanagawa M, Ueda K, Honda O, Tripathi LP, Arai T, Hirose M, Hamasaki T, Yano Y, Kimura T, Kato Y, Takamatsu H, Otsuka T, Minami T, Hirata H, Inoue K, Nagatomo I, Takeda Y, Mori M, Nishikawa H, Mizuguchi K, Kijima T, Kitaichi M, Tomiyama N, Inoue Y, Kumanogoh A. Classification of idiopathic interstitial pneumonias using anti-myxovirus resistance-protein 1 autoantibody. Sci Rep, 7:43201, 2017
12. Mizukami T, Togashi Y, Naruki S, Banno E, Terashima M, de Velasco MA, Sakai K, Yoneshige A, Hayashi H, Fujita Y, Tomida S, Nakajima TE, Fujino T, Boku N, Ito A, Nakagawa K, Nishio K. Significance of FGF9 gene in resistance to anti-EGFR therapies targeting colorectal cancer: A subset of colorectal cancer patients with FGF9 upregulation may be resistant to anti-EGFR therapies. Mol Carcinog, 56:106-117, 2017
13. Kobayashi Y, Azuma K, Nagai H, Kim YH, Togashi Y, Sesumi Y, Chiba M, Shimoji M, Sato K, Tomizawa K, Takemoto T, Nishio K, Mitsudomi T. Characterization of EGFR T790M, L792F, and C797S Mutations as Mechanisms of Acquired Resistance to Afatinib in Lung Cancer. Mol Cancer Ther, 16:357-364, 2017
14. Ueda T, Aokage K, Nishikawa H, Neri S, Nakamura H, Sugano M, Tane K, Miyoshi T, Kojima M, Fujii S, Kuwata T, Ochiai A, Kusumoto M, Suzuki K, Tsuboi M, Ishii G. Immunosuppressive tumor microenvironment of usual interstitial pneumonia-associated squamous cell carcinoma of the lung. J Cancer Res Clin Oncol, 144:835-844, 2018
15. Takeuchi Y, Tanemura A, Tada Y, Katayama I, Kumanogoh A, Nishikawa H. Clinical response to PD-1 blockade correlates with a sub-fraction of peripheral central memory CD4+ T cells in patients with malignant melanoma. Int Immunol, 30:13-22, 2018