Annual Report 2017
Division of Genome Biology
Takashi Kohno, Kouya Shiraishi, Hideaki Ogiwara, Takashi Nakaoku, Kuniko Sunami, Yoko Shimada, Mariko Sasaki, Ayako Otsuka, Maiko Matsuda, Takayuki Honda, Takashi Kuroda, Sou Hirose, Kazushi Yoshida, Tomoko Watanabe, Miyu Yoshida, Yoshie Iga
Our team and what we do
The clarification of somatic mutation signatures in the cancer genome and inter-individual genetic polymorphisms in the human genome is a crucial key to improve cancer medicine. The Division of Genome Biology aims to find "seeds" that are applicable for the development of novel strategies for treatment, diagnosis, and prevention of cancer through identifying and understanding the biological relationship of their seeds with cancer pathogenesis caused by somatic mutations and/or genetic polymorphisms of patients. In order to attain our goal, we are working together with the National Cancer Center (NCC) staff from the NCC Hospital (NCCH), the Exploratory Oncology Research & Clinical Trial Center (EPOC), and the Center for Public Health Sciences to fight lung cancer, the most common cause of cancer-
related death worldwide.
Research activities
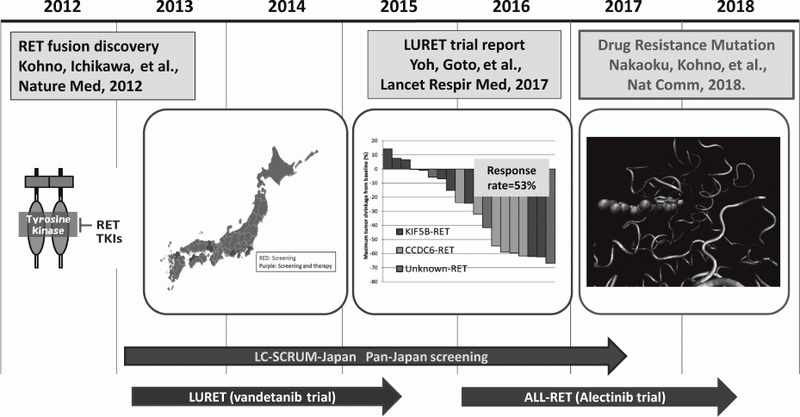
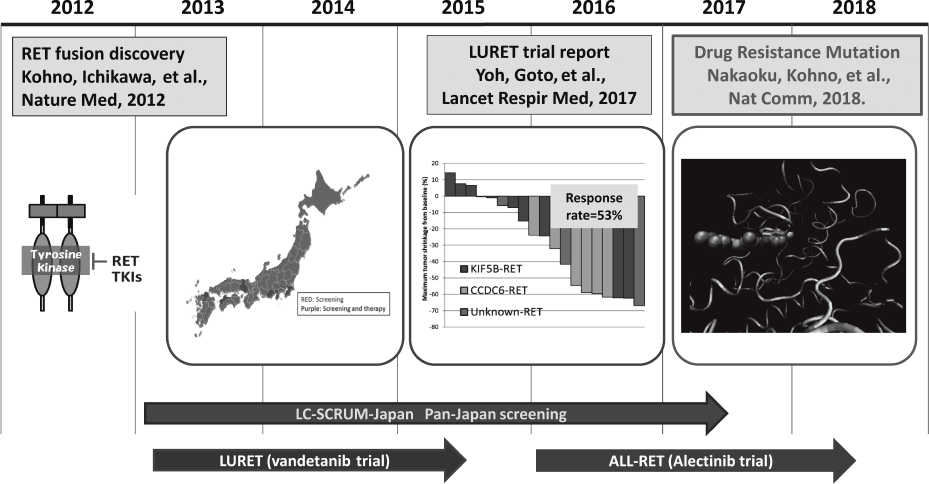
LURET, an open-label, multicenter phase II trial of Vandetanib in patients with RET-
rearranged advanced non-small-cell lung cancer revealed that RET fusion, a new driver oncogene found by us, is a promising therapeutic target. We reported the first case who had of a secondary RET mutation associated with resistance to a type 1 RET tyrosine kinase inhibitor vandetanib. The S904F mutation confers resistance to vandetanib
by increasing the ATP affinity and autophos-
phorylation activity of RET kinase. A reduced interaction with drug by S904F mutation was indicated by thermal shift assay as well as molecular dynamics simulation using K super-computer. Our findings indicate that missense mutations in the activation loop of the kinase domain are able to increase kinase activity and confer drug resistance through allosteric effects.
ARID1A is a gene coding a subunit of SWI/SNF chromatin remodeling complex. ARID1A is frequently mutated in ovarian clear cell carcinoma, ovarian endometrioid carcinoma, uterine corpus endometrial carcinoma, cholangiocarcinoma, and so on. We performed a chemical library
screen to identify synthetic lethal targets for
ARID1A-deficient cancers. We identified a promising drug to specifically kill ARID1A-deficient
cancer cells. An underlying mechanism is being investigated.
Risk factors for lung adeno-carcinogenesis were searched for by genome-wide studies. Genome wide association study (GWAS) of SNPs was conducted as a multi-center collaborative study on 17,106 cases and 158,347 controls. In addition, international GWAS uncovered multiple loci associated with lung cancer risk, such as BPTF, BTNL2, ROS1 and HLA-DPB1. Four major somatic mutational signatures were detected in Japanese LADC genomes, indicating that multiple types of mutation burden, including smoking, causes LADC. To understand mutational and/or genetic features of cancers developed in adolescents and young adults (AYA; 15-39 years old), genome-wide mutational analyses were launched for ovarian, breast, uterine and lung tumors.
Clinical trials
Cases involved in a phase II clinical trial (LURET), which investigates the therapeutic effect of a RET-tyrosine kinase inhibitor, vandetanib, were subjected to a study of drug resistance.
Education
Supervising the research and presentation skills for graduate school students and young researchers.
Future prospects
The goal of our division is the establishment of novel strategies for personalized cancer medicine through the finding of unique "seeds". Genome analyses, including of those of AYA-
cancers, will contribute to precision prevention and therapy of cancers. Understanding biological roles of novel molecular targets, which have been identified by synthetic lethal screen and comprehensive genome analyses, will provide unique and novel concepts improving cancer therapy.
List of papers published in January 2017 - March 2018
Journal
1. Noro R, Ishigame T, Walsh N, Shiraishi K, Robles AI, Ryan BM, Schetter AJ, Bowman ED, Welsh JA, Seike M, Gemma A, Skaug V, Mollerup S, Haugen A, Yokota J, Kohno T, Harris CC. A Two-Gene Prognostic Classifier for Early-Stage Lung Squamous Cell Carcinoma in Multiple Large-Scale and Geographically Diverse Cohorts. J Thorac Oncol, 12:65-76, 2017
2. George J, Saito M, Tsuta K, Iwakawa R, Shiraishi K, Scheel AH, Uchida S, Watanabe SI, Nishikawa R, Noguchi M, Peifer M, Jang SJ, Petersen I, Buttner R, Harris CC, Yokota J, Thomas RK, Kohno T. Genomic Amplification of CD274 (PD-L1) in Small-Cell Lung Cancer. Clin Cancer Res, 23:1220-1226, 2017
3. Yoh K, Seto T, Satouchi M, Nishio M, Yamamoto N, Murakami H, Nogami N, Matsumoto S, Kohno T, Tsuta K, Tsuchihara K, Ishii G, Nomura S, Sato A, Ohtsu A, Ohe Y, Goto K. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med, 5:42-50, 2017
4. Kayama K, Watanabe S, Takafuji T, Tsuji T, Hironaka K, Matsumoto M, Nakayama KI, Enari M, Kohno T, Shiraishi K, Kiyono T, Yoshida K, Sugimoto N, Fujita M. GRWD1 negatively regulates p53 via the RPL11-MDM2 pathway and promotes tumorigenesis. EMBO Rep, 18:123-137, 2017
5. Seow WJ, Matsuo K, Hsiung CA, Shiraishi K, Song M, Kim HN, Wong MP, Hong YC, Hosgood HD 3rd, Wang Z, Chang IS, Wang JC, Chatterjee N, Tucker M, Wei H, Mitsudomi T, Zheng W, Kim JH, Zhou B, Caporaso NE, Albanes D, Shin MH, Chung LP, An SJ, Wang P, Zheng H, Yatabe Y, Zhang XC, Kim YT, Shu XO, Kim YC, Bassig BA, Chang J, Ho JC, Ji BT, Kubo M, Daigo Y, Ito H, Momozawa Y, Ashikawa K, Kamatani Y, Honda T, Sakamoto H, Kunitoh H, Tsuta K, Watanabe SI, Nokihara H, Miyagi Y, Nakayama H, Matsumoto S, Tsuboi M, Goto K, Yin Z, Shi J, Takahashi A, Goto A, Minamiya Y, Shimizu K, Tanaka K, Wu T, Wei F, Wong JY, Matsuda F, Su J, Kim YH, Oh IJ, Song F, Lee VH, Su WC, Chen YM, Chang GC, Chen KY, Huang MS, Yang PC, Lin HC, Xiang YB, Seow A, Park JY, Kweon SS, Chen CJ, Li H, Gao YT, Wu C, Qian B, Lu D, Liu J, Jeon HS, Hsiao CF, Sung JS, Tsai YH, Jung YJ, Guo H, Hu Z, Wang WC, Chung CC, Lawrence C, Burdett L, Yeager M, Jacobs KB, Hutchinson A, Berndt SI, He X, Wu W, Wang J, Li Y, Choi JE, Park KH, Sung SW, Liu L, Kang CH, Hu L, Chen CH, Yang TY, Xu J, Guan P, Tan W, Wang CL, Sihoe AD, Chen Y, Choi YY, Hung JY, Kim JS, Yoon HI, Cai Q, Lin CC, Park IK, Xu P, Dong J, Kim C, He Q, Perng RP, Chen CY, Vermeulen R, Wu J, Lim WY, Chen KC, Chan JK, Chu M, Li YJ, Li J, Chen H, Yu CJ, Jin L, Lo YL, Chen YH, Fraumeni JF Jr, Yamaji T, Yang Y, Hicks B, Wyatt K, Li SA, Dai J, Ma H, Jin G, Song B, Cheng S, Li X, Ren Y, Cui P, Iwasaki M, Shimazu T, Tsugane S, Zhu J, Jiang G, Fei K, Wu G, Chien LH, Chen HL, Su YC, Tsai FY, Chen YS, Yu J, Stevens VL, Laird-Offringa IA, Marconett CN, Lin D, Chen K, Wu YL, Landi MT, Shen H, Rothman N, Kohno T, Chanock SJ, Lan Q. Association between GWAS-identified lung adenocarcinoma susceptibility loci and EGFR mutations in never-smoking Asian women, and comparison with findings from Western populations. Hum Mol Genet, 26:454-465, 2017
6. Ohara K, Arai E, Takahashi Y, Ito N, Shibuya A, Tsuta K, Kushima R, Tsuda H, Ojima H, Fujimoto H, Watanabe SI, Katai H, Kinoshita T, Shibata T, Kohno T, Kanai Y. Genes involved in development and differentiation are commonly methylated in cancers derived from multiple organs: a single-institutional methylome analysis using 1007 tissue specimens. Carcinogenesis, 38:241-251, 2017
7. Kobayashi D, Oike T, Shibata A, Niimi A, Kubota Y, Sakai M, Amornwhichet N, Yoshimoto Y, Hagiwara Y, Kimura Y, Hirota Y, Sato H, Isono M, Yoshida Y, Kohno T, Ohno T, Nakano T. Mitotic catastrophe is a putative mechanism underlying the weak correlation between sensitivity to carbon ions and cisplatin. Sci Rep, 7:40588, 2017
8. Shimada Y, Kohno T, Ueno H, Ino Y, Hayashi H, Nakaoku T, Sakamoto Y, Kondo S, Morizane C, Shimada K, Okusaka T, Hiraoka N. An Oncogenic ALK Fusion and an RRAS Mutation in KRAS Mutation-Negative Pancreatic Ductal Adenocarcinoma. Oncologist, 22:158-164, 2017
9. Miura Y, Inoshita N, Ikeda M, Miyama Y, Oki R, Oka S, Kondoh C, Ozaki Y, Tanabe Y, Kurosawa K, Urakami S, Kohno T, Okaneya T, Takano T. Loss of BAP1 protein expression in the first metastatic site predicts prognosis in patients with clear cell renal cell carcinoma. Urol Oncol, 35:386-391, 2017
10. Sebera J, Hattori Y, Sato D, Reha D, Nencka R, Kohno T, Kojima C, Tanaka Y, Sychrovsky V. The mechanism of the glycosylase reaction with hOGG1 base-excision repair enzyme: concerted effect of Lys249 and Asp268 during excision of 8-oxoguanine. Nucleic Acids Res, 45:5231-5242, 2017
11. Goto A, Tanaka M, Yoshida M, Umakoshi M, Nanjo H, Shiraishi K, Saito M, Kohno T, Kuriyama S, Konno H, Imai K, Saito H, Minamiya Y, Maeda D. The low expression of miR-451 predicts a worse prognosis in non-small cell lung cancer cases. PLoS One, 12:e0181270, 2017
12. Yamamoto Y, Gotoh S, Korogi Y, Seki M, Konishi S, Ikeo S, Sone N, Nagasaki T, Matsumoto H, Muro S, Ito I, Hirai T, Kohno T, Suzuki Y, Mishima M. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat Methods, 14:1097-1106, 2017
13. Saito M, Fujiwara Y, Asao T, Honda T, Shimada Y, Kanai Y, Tsuta K, Kono K, Watanabe S, Ohe Y, Kohno T. The genomic and epigenomic landscape in thymic carcinoma. Carcinogenesis, 38:1084-1091, 2017
14. Lohavanichbutr P, Sakoda LC, Amos CI, Arnold SM, Christiani DC, Davies MPA, Field JK, Haura EB, Hung RJ, Kohno T, Landi MT, Liu G, Liu Y, Marcus MW, O'Kane GM, Schabath MB, Shiraishi K, Slone SA, Tardon A, Yang P, Yoshida K, Zhang R, Zong X, Goodman GE, Weiss NS, Chen C. Common TDP1 Polymorphisms in Relation to Survival among Small Cell Lung Cancer Patients: A Multicenter Study from the International Lung Cancer Consortium. Clin Cancer Res, 23:7550-7557, 2017
15. Sundaresan V, Lin VT, Liang F, Kaye FJ, Kawabata-Iwakawa R, Shiraishi K, Kohno T, Yokota J, Zhou L. Significantly mutated genes and regulatory pathways in SCLC-a meta-analysis. Cancer Genet, 216-217:20-28, 2017
16. Suzuki A, Suzuki M, Mizushima-Sugano J, Frith MC, Makalowski W, Kohno T, Sugano S, Tsuchihara K, Suzuki Y. Sequencing and phasing cancer mutations in lung cancers using a long-read portable sequencer. DNA Res, 24:585-596, 2017
17. Nakaoku T, Kohno T, Araki M, Niho S, Chauhan R, Knowles PP, Tsuchihara K, Matsumoto S, Shimada Y, Mimaki S, Ishii G, Ichikawa H, Nagatoishi S, Tsumoto K, Okuno Y, Yoh K, McDonald NQ, Goto K. A secondary RET mutation in the activation loop conferring resistance to vandetanib. Nat Commun, 9:625, 2018
18. Saito M, Saito K, Shiraishi K, Maeda D, Suzuki H, Minamiya Y, Kono K, Kohno T, Goto A. Identification of candidate responders for anti-PD-L1/PD-1 immunotherapy, Rova-T therapy, or EZH2 inhibitory therapy in small-cell lung cancer. Mol Clin Oncol, 8:310-314, 2018
19. Kanke Y, Shimomura A, Saito M, Honda T, Shiraishi K, Shimada Y, Watanabe R, Yoshida H, Yoshida M, Shimizu C, Takahashi K, Totsuka H, Ogiwara H, Hirose S, Kono K, Tamura K, Okamoto A, Kinoshita T, Kato T, Kohno T. Gene aberration profile of tumors of adolescent and young adult females. Oncotarget, 9:6228-6237, 2018
20. Sereewattanawoot S, Suzuki A, Seki M, Sakamoto Y, Kohno T, Sugano S, Tsuchihara K, Suzuki Y. Identification of potential regulatory mutations using multi-omics analysis and haplotyping of lung adenocarcinoma cell lines. Sci Rep, 8:4926, 2018
21. Saito M, Suzuki H, Kono K, Takenoshita S, Kohno T. Treatment of lung adenocarcinoma by molecular-targeted therapy and immunotherapy. Surg Today, 48:1-8, 2018
22. Kohno T. Implementation of "clinical sequencing" in cancer genome medicine in Japan. Cancer Sci, 109:507-512, 2018
