Annual Report 2018
Division of Molecular Modification and Cancer Biology
Ryuji Hamamoto, Syuzo Kaneko, Masaaki Komatsu, Ken Asada, Masayoshi Yamada, Masamichi Takahashi, Kazuma Kobayashi, Kyoko Fujioka, Noriko Ikawa, Hiroko Kondo, Shigemi Yamada, Kruthi Suvarna, Hidenori Machino, Amina Bolatkan, Norio Shinkai, Kanto Syozu, Ai Dozen, Ryota Mieda
Introduction
In our division, we try to elucidate the molecular mechanism of how abnormal modification causes human tumorigenesis, in particular focusing on protein methylation, and apply our findings for development of novel anti-cancer drugs. In addition, we introduced AI technologies into a medical study during the early stages of the development of medical AI in Japan and developed an integrated cancer medical system using AI technology with the intention of applying it to Precision Medicine. We now play a central role in the medical AI research field in Japan.
Research activities
1. Development of a next-generation ChIP-seq method with the intention of applying it to Precision Medicine.
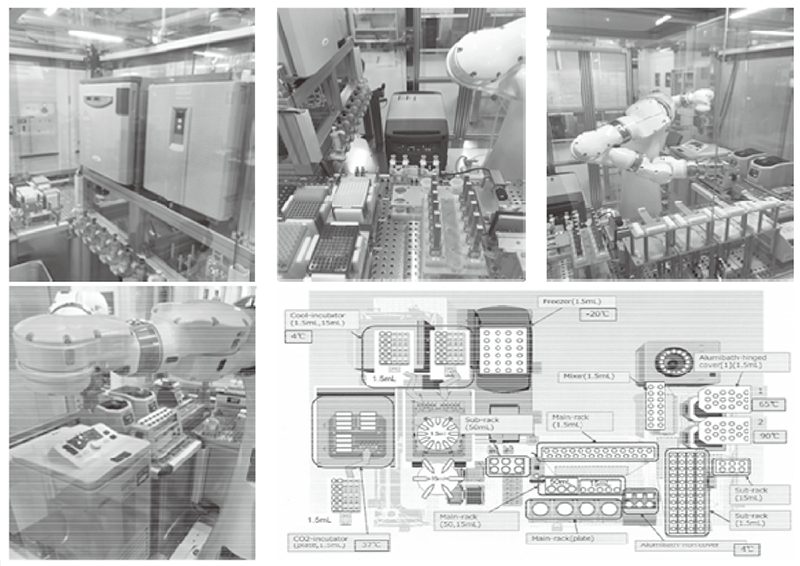
In order to establish the platform to conduct multi-omics analysis using machine learning and deep learning, we tried to develop a nextgeneration ChIP-seq method. Consequently, we succeeded in developing a system that enables the ChIP-seq analysis of formalin-fixed, paraffinembedded (FFPE) samples effectively. On the basis of this system, we successfully identified not only the status of histone modifications (H3K27 acetylation, H3K4 tri-methylation) but also the identification of the binding sites of transcription factor CCCTC-binding Factor (CTCF). This is the first case to succeed in identifying the transcription factor binding sites by ChIP-seq analysis using the FFPE sample. In this system, we introduced the human general-purpose robot LabDroid, which enabled acquisition of highly robust data (Figure 1).
Figure 1. Automation system of next-generation ChIP-seq with the bio experiment automation robot

2. Development of highly precise image diagnosis devices and image diagnosis methods by AI technology.
As for the endoscope image analysis, we developed a system that immediately detects colorectal cancer and ulcerative colon polyps, a precursor to cancer, during an endoscopic examination using artificial intelligence (AI). It automatically detects colorectal cancer and polyps from images and videos taken during an endoscopic examination of the colon, and aids in discovery of lesions by endoscopists. It improves polyp detection, which was an issue during such exams, and increases the detection rate. In this manner, it greatly contributes to the prevention and early detection of colorectal cancer. In addition, we built an annotation platform designed from the viewpoint of the clinician, and designed an integrated cancer medical care database. The results have already been installed in our partner company to achieve their social implementation
3. Construction of multi-omics analysis systems using artificial intelligence technologies
In this study, we started developing an integrated lung cancer database including whole exome sequencing data, transcriptome analysis data, histone modification data, DNA methylation data and radiation image data, and developed an analysis algorithm.
Education
Seven graduate school students from the University of Tokyo, Tokyo Medical and Dental University, Juntendo University, Toyama University and Keio University were trained in our revision. One member in the division won the NCCRI director award.
Future prospects
1) To develop an analytical method of multiomics data using AI technology to apply to Precision Medicine.
2) To analyze the chromatin structure of clinical tissues using ATAC-seq and ChIPseq methods, and examine the possibility of clinical application.
3) To develop an integrated medical system for cancer treatment using AI technology.
List of papers published in 2018
Journal
1. Toyokawa G, Takada K, Tagawa T, Hamamoto R, Yamada Y, Shimokawa M, Oda Y, Maehara Y. A Positive Correlation Between the EZH2 and PD-L1 Expression in Resected Lung Adenocarcinomas. Ann Thorac Surg, 107:393-400, 2019
2. Kim SK, Kim K, Ryu JW, Ryu TY, Lim JH, Oh JH, Min JK, Jung CR, Hamamoto R, Son MY, Kim DS, Cho HS. The novel prognostic marker, EHMT2, is involved in cell proliferation via HSPD1 regulation in breast cancer. Int J Oncol, 54:65-76, 2019
3. Kukita A, Sone K, Oda K, Hamamoto R, Kaneko S, Komatsu M, Wada M, Honjoh H, Kawata Y, Kojima M, Oki S, Sato M, Asada K, Taguchi A, Miyasaka A, Tanikawa M, Nagasaka K, Matsumoto Y, Wada-Hiraike O, Osuga Y, Fujii T. Histone methyltransferase SMYD2 selective inhibitor LLY-507 in combination with poly ADP ribose polymerase inhibitor has therapeutic potential against high-grade serous ovarian carcinomas. Biochem Biophys Res Commun, 513:340-346, 2019
4. Piao L, Fujioka K, Nakakido M, Hamamoto R. Regulation of poly(ADP-Ribose) polymerase 1 functions by post-translational modifications. Front Biosci (Landmark Ed), 23:13-26, 2018
5. Kunizaki M, Tominaga T, Wakata K, Miyazaki T, Matsumoto K, Sumida Y, Hidaka S, Yamasaki T, Yasutake T, Sawai T, Hamamoto R, Nanashima A, Nagayasu T. Clinical significance of the C-reactive protein-to-albumin ratio for the prognosis of patients with esophageal squamous cell carcinoma. Mol Clin Oncol, 8:370-374, 2018
6. Toyokawa G, Takada K, Tagawa T, Kinoshita F, Kozuma Y, Matsubara T, Haratake N, Takamori S, Akamine T, Hirai F, Yamada Y, Hamamoto R, Oda Y, Maehara Y. Prevalence of Enhancer of Zeste Homolog 2 in Patients with Resected Small Cell Lung Cancer. Anticancer Res, 38:3707-3711, 2018
7. Shigekawa Y, Hayami S, Ueno M, Miyamoto A, Suzaki N, Kawai M, Hirono S, Okada KI, Hamamoto R, Yamaue H. Overexpression of KDM5B/JARID1B is associated with poor prognosis in hepatocellular carcinoma. Oncotarget, 9:34320-34335, 2018
8. Lee CH, Yu JR, Kumar S, Jin Y, LeRoy G, Bhanu N, Kaneko S, Garcia BA, Hamilton AD, Reinberg D. Allosteric Activation Dictates PRC2 Activity Independent of Its Recruitment to Chromatin. Mol Cell, 70:422-434.e6, 2018
