Annual Report 2019
Division of Cancer Genomics
Tatsuhiro Shibata, Yasushi Totoki, Yasuhito Arai, Natsuko Hama, Hiromi Nakamura, Fumie Hosoda, Hirofumi Rokutan, Mihoko Adachi, Hiroshi Chikuta, Akihiko Fukagawa, Yuuya Kobayashi, Wakako Mukai, Erika Arakawa, Seri Yamagishi, Hiroe Nozaki, Machiko Watanabe
Introduction
The Division of Cancer Genomics focuses on comprehensive characterization of the cancer genome on the basis of tumor pathology and aims to make a “breakthrough” by identifying novel cancer-related genes, including potential therapeutic targets and biomarkers, and to understand the cancer genome as heterogeneous but intervention-able “biological systems” that contribute to the pathogenesis of cancer. This Division has also been participating in an international consortium (International Cancer Genome Consortium (ICGC) and ICGC-ARGO), contributed to the core facility of the center, and has been developing new informatics tools for the data analysis from various types of next-generation high-performance sequencers.
Research activities
We participated in the Pan-Cancer Analysis of Whole Genomes Consortium of the ICGC and reported the integrated analysis of 2,658 cancer whole-genome data to uncover the panorama of cancer drivers including non-coding genomes.
We performed an integrated genomic and transcriptomic analysis of the largest multi-ethnic gastric cancer cohort and identified more than 100 highly mutated genes that are statistically significant and selectively mutated, and established a driver gene landscape of gastric cancer containing many new ones. Using a combination of whole-genome sequencing (WGS), RNA sequencing (RNAseq), assay for transposase-accessible chromatin using sequencing and enzymatic methyl sequencing, we have analyzed the relation between genetic aberrations and epigenetic features, and mutational signatures characterized among ethnic groups in kidney cancers. To identify the genomic landscape and therapeutic targets in esophageal squamous cell carcinoma (ESCC), we performed an integrated analysis WGS and RNAseq of ESCC cases.
For the development of a novel molecular target therapy, we have explored molecular detection and clinicopathological characteristics of advanced/recurrent biliary tract carcinomas (BTC) harboring the FGFR2 rearrangements in a prospective multicenter study (PRELUDE). We developed FISH assay and RNA sequencing analysis systems to detect FGFR2 rearrangements using FFPE specimens from small biopsy samples including EUS-FNA (Figure 1). Clinical trials for BTC patients with FGFR2 rearrangements are in progress.
Figure 1.Detection of FGFR2 fusion in a cholangiocarcinoma case by FISH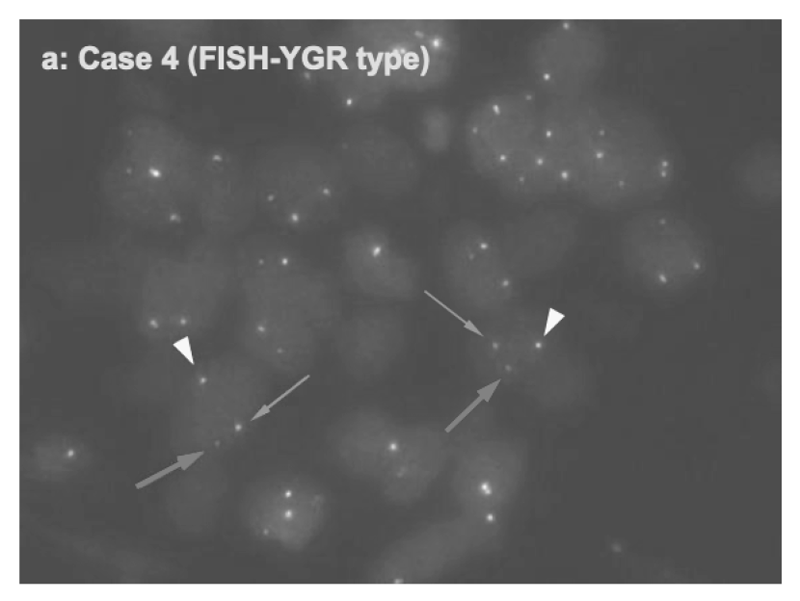

We performed fecal metagenomic and metabolomic studies on samples from a large cohort of 616 participants, and identified that shifts in the microbiome and metabolome occur from the very early stages of the development of colorectal cancer, which is of possible etiological and diagnostic importance (Figure 2).
Figure 2.Metagenomic and metabolomic changes during colon carcinogenesis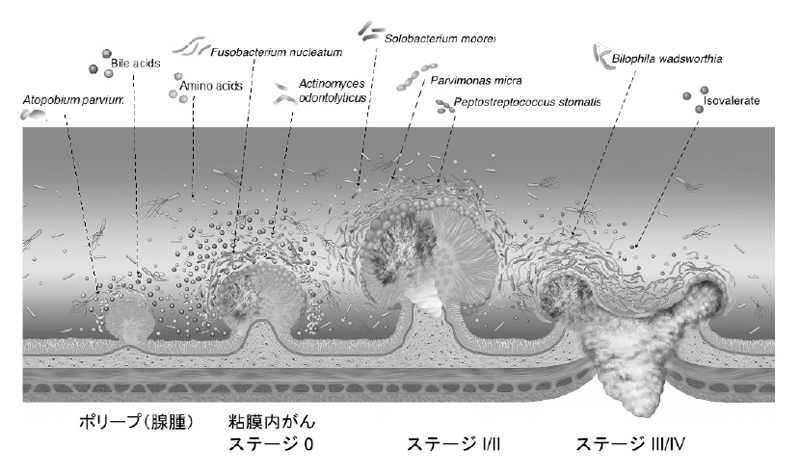

A fraction of sarcomas remains unclassified or difficult to classify based on the current histologic and immunohistochemical profile. In the process of our systematic sequencing project for these unclassifiable sarcomas, we identified driver fusions involving the KMT2A (MLL) gene. The fusions account for a subset of aggressive unclassifiable sarcomas in young adults (AYA). We also uncovered unique molecular entities including BCOR-associated sarcoma, MGA-NUTM1 sarcoma, and H3F3A-negative giant cell tumor.
Future prospects
By utilizing current and cutting-edge sequencing technologies (e.g. long-read and single-cell sequencing), this division will actively investigate the cancer genomics from both basic (new biomarkers including therapeutic targets, epigenomics, metagenomics and immune-genomics) and translational research (preclinical research, liquid clinical sequencing, PGx and germline evaluation) viewpoints. Especially, extensive collaboration with cancer-immunology groups by applying single cell immune-profiling and TCR repertoire sequencing will be achieved. This division will also contribute to the development of bioinformatics tools and human resources for analyzing the large cancer genomics data.
List of papers published in 2019
Journal
1. Mimaki S, Watanabe M, Kinoshita M, Yamashita R, Haeno H, Takemura S, Tanaka S, Marubashi S, Totsuka Y, Shibata T, Nakagama H, Ochiai A, Nakamori S, Kubo S, Tsuchihara K. Multifocal origin of occupational cholangiocarcinoma revealed by comparison of multilesion mutational profiles. Carcinogenesis, 41:368-376, 2020
2. ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature, 578:82-93, 2020
3. Takai E, Maeda D, Li Z, Kudo-Asabe Y, Totoki Y, Nakamura H, Nakamura A, Nakamura R, Kirikawa M, Ito Y, Yoshida M, Inoue T, Habuchi T, Ikoma S, Katoh H, Kato M, Shibata T, Ishikawa S, Yachida S, Goto A. Post-mortem Plasma Cell-Free DNA Sequencing: Proof-of-Concept Study for the “Liquid Autopsy”. Sci Rep, 10:2120, 2020
4. Goto T, Arai Y, Shibata T, Oyama T, Yoshida A. Sarcoma with MGA-NUTM1 fusion in the lung: an emerging entity. Virchows Arch, 476:317-322, 2020
5. Yoshida A, Arai Y, Hama N, Chikuta H, Bando Y, Nakano S, Kobayashi E, Shibahara J, Fukuhara H, Komiyama M, Watanabe SI, Tamura K, Kawai A, Shibata T. Expanding the clinicopathologic and molecular spectrum of BCOR-associated sarcomas in adults. Histopathology, 76:509-520, 2020
6. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, Islam SMA, Lopez-Bigas N, Klimczak LJ, McPherson JR, Morganella S, Sabarinathan R, Wheeler DA, Mustonen V, Getz G, Rozen SG, Stratton MR. The repertoire of mutational signatures in human cancer. Nature, 578:94-101, 2020
7. Jiao W, Atwal G, Polak P, Karlic R, Cuppen E, Danyi A, de Ridder J, van Herpen C, Lolkema MP, Steeghs N, Getz G, Morris Q, Stein LD. A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat Commun, 11:728, 2020
8. Totsuka Y, Lin Y, He Y, Ishino K, Sato H, Kato M, Nagai M, Elzawahry A, Totoki Y, Nakamura H, Hosoda F, Shibata T, Matsuda T, Matsushima Y, Song G, Meng F, Li D, Liu J, Qiao Y, Wei W, Inoue M, Kikuchi S, Nakagama H, Shan B. DNA Adductome Analysis Identifies N-Nitrosopiperidine Involved in the Etiology of Esophageal Cancer in Cixian, China. Chem Res Toxicol, 32:1515-1527, 2019
9. Hirata M, Asano N, Katayama K, Yoshida A, Tsuda Y, Sekimizu M, Mitani S, Kobayashi E, Komiyama M, Fujimoto H, Goto T, Iwamoto Y, Naka N, Iwata S, Nishida Y, Hiruma T, Hiraga H, Kawano H, Motoi T, Oda Y, Matsubara D, Fujita M, Shibata T, Nakagawa H, Nakayama R, Kondo T, Imoto S, Miyano S, Kawai A, Yamaguchi R, Ichikawa H, Matsuda K. Integrated exome and RNA sequencing of dedifferentiated liposarcoma. Nat Commun, 10:5683, 2019
10. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med, 25:968-976, 2019
11. Yoshida A, Arai Y, Tanzawa Y, Wakai S, Hama N, Kawai A, Shibata T. KMT2A (MLL) fusions in aggressive sarcomas in young adults. Histopathology, 75:508-516, 2019
12. Yoshida KI, Nakano Y, Honda-Kitahara M, Wakai S, Motoi T, Ogura K, Sano N, Shibata T, Okuma T, Iwata S, Kawai A, Ichimura K, Yoshida A. Absence of H3F3A mutation in a subset of malignant giant cell tumor of bone. Mod Pathol, 32:1751-1761, 2019
13. Lee JS, Rhee TM, Pietrasz D, Bachet JB, Laurent-Puig P, Kong SY, Takai E, Yachida S, Shibata T, Lee JW, Park HC, Zang DY, Jeon K, Lee J, Kim M, Kim HS, Kang HJ, Lee YK. Circulating tumor DNA as a prognostic indicator in resectable pancreatic ductal adenocarcinoma: A systematic review and meta-analysis. Sci Rep, 9:16971, 2019
14. Hayashi Y, Goyama S, Liu X, Tamura M, Asada S, Tanaka Y, Fukuyama T, Wunderlich M, O'Brien E, Mizukawa B, Yamazaki S, Matsumoto A, Yamasaki S, Shibata T, Matsuda K, Sashida G, Takizawa H, Kitamura T. Antitumor immunity augments the therapeutic effects of p53 activation on acute myeloid leukemia. Nat Commun, 10:4869, 2019
15. Kikuchi-Koike R, Nagasaka K, Tsuda H, Ishii Y, Sakamoto M, Kikuchi Y, Fukui S, Miyagawa Y, Hiraike H, Kobayashi T, Kinoshita T, Kanai Y, Shibata T, Imoto I, Inazawa J, Matsubara O, Ayabe T. Array comparative genomic hybridization analysis discloses chromosome copy number alterations as indicators of patient outcome in lymph node-negative breast cancer. BMC Cancer, 19:521, 2019
16. Tsuda Y, Hirata M, Katayama K, Motoi T, Matsubara D, Oda Y, Fujita M, Kobayashi H, Kawano H, Nishida Y, Sakai T, Okuma T, Goto T, Ogura K, Kawai A, Ae K, Anazawa U, Suehara Y, Iwata S, Miyano S, Imoto S, Shibata T, Nakagawa H, Yamaguchi R, Tanaka S, Matsuda K. Massively parallel sequencing of tenosynovial giant cell tumors reveals novel CSF1 fusion transcripts and novel somatic CBL mutations. Int J Cancer, 145:3276-3284, 2019
17. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, Gandini S, Serrano D, Tarallo S, Francavilla A, Gallo G, Trompetto M, Ferrero G, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Wirbel J, Schrotz-King P, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G, Cordero F, Dias-Neto E, Setubal JC, Tett A, Pardini B, Rescigno M, Waldron L, Naccarati A, Segata N. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med, 25:667-678, 2019
18. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, Sunagawa S, Coelho LP, Schrotz-King P, Vogtmann E, Habermann N, Niméus E, Thomas AM, Manghi P, Gandini S, Serrano D, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Waldron L, Naccarati A, Segata N, Sinha R, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med, 25:679-689, 2019
19. Rokutan H, Abe H, Nakamura H, Ushiku T, Arakawa E, Hosoda F, Yachida S, Tsuji Y, Fujishiro M, Koike K, Totoki Y, Fukayama M, Shibata T. Initial and crucial genetic events in intestinal-type gastric intramucosal neoplasia. J Pathol, 247:494-504, 2019
