Annual Report 2019
Division of Molecular Modification and Cancer Biology
Ryuji Hamamoto, Syuzo Kaneko, Masaaki Komatsu, Ken Asada, Satoshi Takahashi, Ken Takasawa, Masayoshi Yamada, Masamichi Takahashi, Kazuma Kobayashi, Kyoko Fujioka, Noriko Ikawa, Hiroko Kondo, Shigemi Yamada, Kruthi Suvarna, Hidenori Machino, Amina Bolatkan, Norio Shinkai, Kanto Shozu, Ai Dozen, Ryota Mieda
Introduction
In our division, we analyze the abnormal molecular modifications in tumors, especially focusing on histone modifications, and pioneer the introduction of AI technology into medical research in Japan to develop an integrated cancer treatment system. In particular, as the core group of the PRISM project supervised by the Cabinet Office, we play a central role in promoting medical AI research in Japan.
Research activities
1. Development of the RCRA ChIP-seq method for application to precision medicine
We have developed the RCRA ChIP-seq method as a basis for multi-layered omics analysis. As a result, we succeeded in developing a protocol for efficient ChIP-seq analysis from FFPE tissues. By using this method, we succeeded in identifying the binding site of transcription factors in a way that was unprecedented anywhere in the world. In this method, robotics was also introduced to enable reproducible and accurate data acquisition.
2. Development of a high-precision image diagnosis system using AI technology
With regard to the development of the real-time endoscopy diagnostic support system using AI technology, we presented a paper and conducted clinical trials. On the basis of the results, we are currently submitting an application to the PMDA for regulatory approval. In addition, we have developed an AI-based platform for automatic annotation of radiological images, which has already been out-licensed to a major medical device manufacturer after receiving a patent.
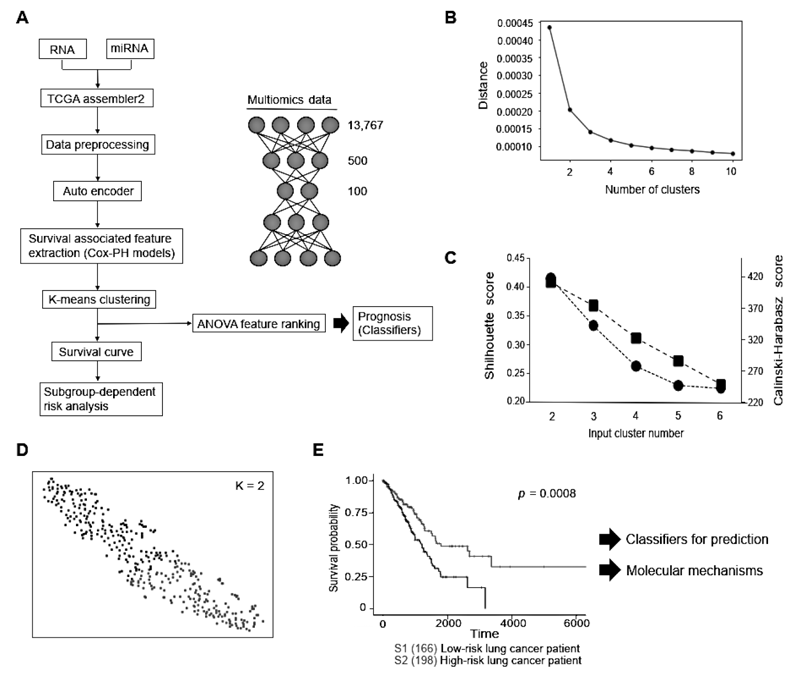
3. Construction of the world's largest integrated lung cancer database and development of multi-omics analysis platforms using AI technology
We have succeeded in constructing one of the world's largest integrated lung cancer database with more than 1500 cases of lung cancer. In addition, we have developed an efficient system to collect medical information and medical imaging information of various modalities from electronic medical records and pathology systems. Furthermore, we have developed several platforms based on AI technology to efficiently and accurately analyze large-scale omics data on cancer. The results of our analysis platform using an autoencoder are shown in Figure 1.
Education
Six graduate school students from the University of Tokyo, Tokyo Medical and Dental University, Juntendo University, Toyama University and Keio University were trained in our division. One member in the division won the NCCRI director award.
Future prospects
1) We will conduct ChIP-seq analysis of a large number of lung cancer samples using RCRA ChIP-seq.
2) As for medical image analysis using AI technology, we will present papers on this subject and aim to implement our achievements in society.
3) We aim to develop new cancer diagnostics and treatments by building platforms for omics analysis using machine learning and deep learning technologies to elucidate molecular mechanisms of cancer.
List of papers published in 2019
Journal
1. Hirose S, Murakami N, Takahashi K, Kuno I, Takayanagi D, Asami Y, Matsuda M, Shimada Y, Yamano S, Sunami K, Yoshida K, Honda T, Nakahara T, Watanabe T, Komatsu M, Hamamoto R, Kato MK, Matsumoto K, Okuma K, Kuroda T, Okamoto A, Itami J, Kohno T, Kato T, Shiraishi K, Yoshida H. Genomic alterations in STK11 can predict clinical outcomes in cervical cancer patients. Gynecol Oncol, 156:203-210, 2020
2. Asada K, Kobayashi K, Joutard S, Tubaki M, Takahashi S, Takasawa K, Komatsu M, Kaneko S, Sese J, Hamamoto R. Uncovering Prognosis-Related Genes and Pathways by Multi-Omics Analysis in Lung Cancer. Biomolecules, 10:524, 2020
3. Isago H, Mitani A, Mikami Y, Horie M, Urushiyama H, Hamamoto R, Terasaki Y, Nagase T. Epithelial Expression of YAP and TAZ Is Sequentially Required in Lung Development. Am J Respir Cell Mol Biol, 62:256-266, 2020
4. Matsuno Y, Atsumi Y, Shimizu A, Katayama K, Fujimori H, Hyodo M, Minakawa Y, Nakatsu Y, Kaneko S, Hamamoto R, Shimamura T, Miyano S, Tsuzuki T, Hanaoka F, Yoshioka KI. Replication stress triggers microsatellite destabilization and hypermutation leading to clonal expansion in vitro. Nat Commun, 10:3925, 2019
5. Hamamoto R, Komatsu M, Takasawa K, Asada K, Kaneko S. Epigenetics Analysis and Integrated Analysis of Multiomics Data, Including Epigenetic Data, Using Artificial Intelligence in the Era of Precision Medicine. Biomolecules, 10:62, 2019
6. Kim S, Bolatkan A, Kaneko S, Ikawa N, Asada K, Komatsu M, Hayami S, Ojima H, Abe N, Yamaue H, Hamamoto R. Deregulation of the Histone Lysine-Specific Demethylase 1 Is Involved in Human Hepatocellular Carcinoma. Biomolecules, 9:810, 2019
7. Kobayashi K, Murakami N, Takahashi K, Inaba K, Igaki H, Hamamoto R, Itami J. A Population-based Statistical Model for Investigating Heterogeneous Intraprostatic Sensitivity to Radiation Toxicity After (125)I Seed Implantation. In Vivo, 33:2103-2111, 2019
8. Yamada M, Saito Y, Imaoka H, Saiko M, Yamada S, Kondo H, Takamaru H, Sakamoto T, Sese J, Kuchiba A, Shibata T, Hamamoto R. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci Rep, 9:14465, 2019
9. Kobayashi K, Murakami N, Takahashi K, Inaba K, Hamamoto R, Itami J. Local Radiotherapy or Chemotherapy for Oligo-recurrent Cervical Cancer in Patients With Prior Pelvic Irradiation. In Vivo, 33:1659-1665, 2019
10. Kojima M, Sone K, Oda K, Hamamoto R, Kaneko S, Oki S, Kukita A, Machino H, Honjoh H, Kawata Y, Kashiyama T, Asada K, Tanikawa M, Mori-Uchino M, Tsuruga T, Nagasaka K, Matsumoto Y, Wada-Hiraike O, Osuga Y, Fujii T. The histone methyltransferase WHSC1 is regulated by EZH2 and is important for ovarian clear cell carcinoma cell proliferation. BMC Cancer, 19:455, 2019
11. Kukita A, Sone K, Oda K, Hamamoto R, Kaneko S, Komatsu M, Wada M, Honjoh H, Kawata Y, Kojima M, Oki S, Sato M, Asada K, Taguchi A, Miyasaka A, Tanikawa M, Nagasaka K, Matsumoto Y, Wada-Hiraike O, Osuga Y, Fujii T. Histone methyltransferase SMYD2 selective inhibitor LLY-507 in combination with poly ADP ribose polymerase inhibitor has therapeutic potential against high-grade serous ovarian carcinomas. Biochem Biophys Res Commun, 513:340-346, 2019
Book
1. Yasutomi S, Arakaki T, Hamamoto R. Shadow Detection for Ultrasound Images Using Unlabeled Data and Synthetic Shadows. In: arXiv preprint, pp arXiv:1908.01439, 2019
