Annual Report 2021
Division of Epigenomics
Toshikazu Ushijima, Satoshi Yamashita, Hideyuki Takeshima, Naoko Hattori, Harumi Yamada, Yuko Miyaji, Mika Wakabayashi, Naoko Takagi, Kazuhiro Nishiyama, Chihiro Takeuchi, Yuyu Liu, Sho Ueda, Chun-Dong Zhang, Takahiro Ebata, Yoshimi Yasukawa, Nobuaki Arai, Gina Miku Oba
Introduction
This Division has been focusing on the epigenetic mechanisms of carcinogenesis, and has identified many aberrantly methylated genes in various cancers, including gastric cancers, esophageal squamous cell carcinomas (ESCCs), neuroblastomas, breast cancers, pancreatic cancers, lung cancers, ovarian cancers, and melanomas. These findings have led to identification of novel tumor-suppressor genes, development of powerful biomarkers, and establishment of the concept of an "epigenetic field for cancerization (epigenetic field defect)". This Division 1) has revealed induction mechanisms of epigenetic alterations, 2) has developed clinically useful biomarkers, and 3) has been engaged in development of an epigenetic cancer therapy.
Research activities
1. Induction Mechanisms of Epigenetic Alterations
Induction mechanisms of epigenetic alterations are important to understand cancers. This year, it was revealed that mutations of ARID1A, a component of chromatin remodeling complex SWI/SNF, were associated with the CpG island methylator phenotype (CIMP) in gastric cancers. Mechanistically, ARID1A depletion had a causal role in aberrant DNA methylation induction.
2. Development of Biomarkers
This Division previously conducted a multicenter prospective cohort study for the prediction of metachronous gastric cancer risk after endoscopic resection, and showed that "the measurement of aberrant DNA methylation accumulated in normal tissues is clinically useful for cancer risk diagnosis". Based on this result, we have been conducting a multicenter prospective cohort study for the prediction of gastric cancer risk in healthy volunteers who underwent eradication of Helicobacter pylori, the almost exclusive cause of gastric cancers. This year, we observed the development of gastric cancers in 25 of 1,880 recruited people with a median follow-up of 3.14 years. We will continue the follow-up, and perform the statistical analysis when the median follow-up period reaches 5 years.
Somatic mutations, in addition to aberrant DNA methylation, accumulated in normal human tissues are useful for cancer risk diagnosis. This year, we developed a novel sequencing method [enzymatically cleaved and optimal sequencing (EcoSeq)], which can count somatic mutations in a single DNA molecule with a sensitivity of as low as 3 × 10-8 per base pair (bp) (Figure 1). Using this method, the mutation frequency in normal peripheral blood cells of pediatric sarcoma patients who received chemotherapy was shown to be increased (Figure 2).
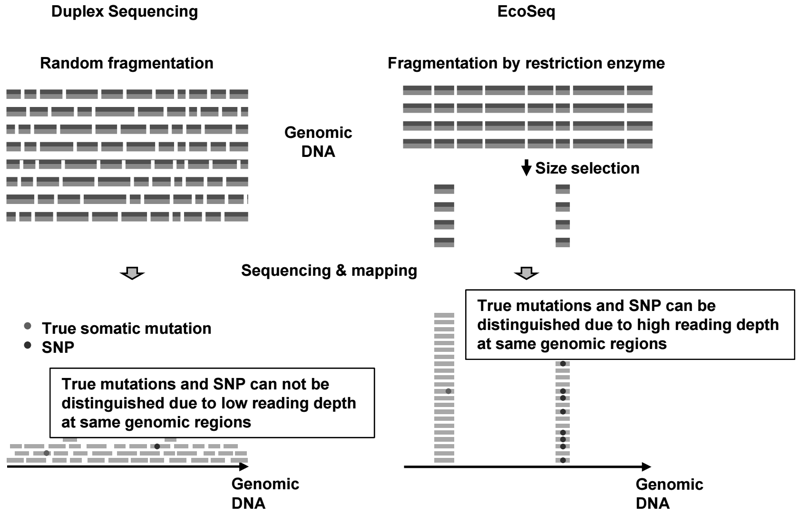
Figure 2. Increase of somatic mutations in normal peripheral
blood cells by chemotherapy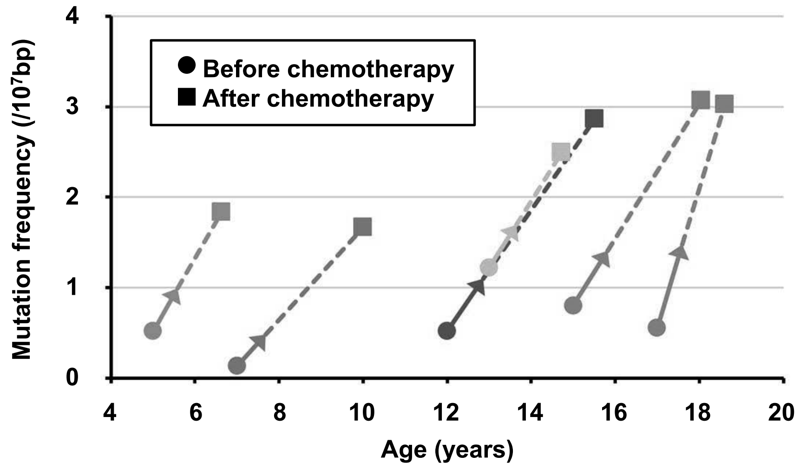

3. Development of epigenetic cancer therapy
DNA demethylating therapy with low-dose prolonged drug exposure shows a remarkable therapeutic efficacy. This year, it was revealed that the combination of tamibarotene (a synthetic retinoid) and a decitabine (DNA demethylating agent) induced differentiation of neuroblastomas through retinoic acid signal reprogramming.
Education
We have accepted five trainees from Shonan Kamakura General Hospital, University of Tokyo, Yokohama City University, Juntendo University, and Kagawa University.
Future Prospects
Based on these results, we will 1) continue multicenter prospective cohort studies for the prediction of gastric cancer risk, and 2) conduct the development of epigenetic cancer prevention and therapy.
List of papers published in 2021
Journal
1. Zhang CD, Takeshima H, Sekine S, Yamashita S, Liu YY, Hattori N, Abe H, Yamashita H, Fukuda M, Imamura Y, Ushiku T, Katai H, Makino H, Watanabe M, Seto Y, Ushijima T. Prediction of tissue origin of adenocarcinomas in the esophagogastric junction by DNA methylation. Gastric Cancer, 25:336-345, 2022
2. Ebata T, Yamashita S, Takeshima H, Yoshida H, Kawata Y, Kino N, Yasugi T, Terao Y, Yonemori K, Kato T, Ushijima T. DNA methylation marker to estimate ovarian cancer cell fraction. Med Oncol, 39:78, 2022
3. Yamada H, Takeshima H, Fujiki R, Yamashita S, Sekine S, Ando T, Hattori N, Okabe A, Yoshikawa T, Obama K, Katai H, Kaneda A, Ushijima T. ARID1A loss-of-function induces CpG island methylator phenotype. Cancer letters, 532:215587, 2022
4. Takeuchi C, Sato J, Yamashita S, Sasaki A, Akahane T, Aoki R, Yamamichi M, Liu YY, Ito M, Furuta T, Nakajima S, Sakaguchi Y, Takahashi Y, Tsuji Y, Niimi K, Tomida S, Fujishiro M, Yamamichi N, Ushijima T. Autoimmune gastritis induces aberrant DNA methylation reflecting its carcinogenic potential.. Journal of gastroenterology, 57:144-155, 2022
5. Kamachi K, Ureshino H, Watanabe T, Yoshida N, Yamamoto Y, Kurahashi Y, Fukuda-Kurahashi Y, Hayashi Y, Hirai H, Yamashita S, Ushijima T, Okada S, Kimura S. Targeting DNMT1 by demethylating agent OR-2100 increases tyrosine kinase inhibitors-sensitivity and depletes leukemic stem cells in chronic myeloid leukemia.. Cancer letters, 526:273-283, 2022
6. Satomi K, Takami H, Fukushima S, Yamashita S, Matsushita Y, Nakazato Y, Suzuki T, Tanaka S, Mukasa A, Saito N, Kanamori M, Kumabe T, Tominaga T, Kobayashi K, Nagane M, Iuchi T, Yoshimoto K, Tamura K, Maehara T, Sakai K, Sugiyama K, Yokogami K, Takeshima H, Nonaka M, Asai A, Ushijima T, Matsutani M, Nishikawa R, Ichimura K. 12p gain is predominantly observed in non-germinomatous germ cell tumors and identifies an unfavorable subgroup of central nervous system germ cell tumors.. Neuro-oncology, 24:834-846, 2022
7. Motoo I, Nanjo S, Ando T, Yamashita S, Ushijima T, Yasuda I. Methylation silencing of ULK2 via epithelial-mesenchymal transition causes transformation to poorly differentiated gastric cancers.. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association, 25:325-335, 2022
8. Nitani C, Hara J, Kawamoto H, Taguchi T, Kimura T, Yoshimura K, Hamada A, Kitano S, Hattori N, Ushijima T, Ono H, Nakamoto M, Higuchi T, Sato A. Phase I study of tamibarotene monotherapy in pediatric and young adult patients with recurrent/refractory solid tumors. Cancer Chemother Pharmacol, 88:99-107, 2021
9. Ureshino H, Kurahashi Y, Watanabe T, Yamashita S, Kamachi K, Yamamoto Y, Fukuda-Kurahashi Y, Yoshida-Sakai N, Hattori N, Hayashi Y, Kawaguchi A, Tohyama K, Okada S, Harada H, Ushijima T, Kimura S. Silylation of deoxynucleotide analog yields an orally available drug with anti-leukemia effects. Mol Cancer Ther, 20:1412-1421, 2021
10. Katsuda Y, Tanaka K, Mori T, Narita M, Takeshima H, Kondo T, Yamabe Y, Matsufuji M, Sato D, Hamada Y, Yamaguchi K, Ushijima T, Inada E, Kuzumaki N, Iseki M, Narita M. Histone modification of pain-related gene expression in spinal cord neurons under a persistent postsurgical pain-like state by electrocautery. Mol Brain, 14:146, 2021
11. Ueda S, Yamashita S, Watanabe SI, Wakabayashi M, Motoi N, Noguchi M, Sekine S, Sato Y, Ushijima T. Influence of degree of DNA degradation in formalin-fixed and paraffin-embedded tissue samples on accuracy of genome-wide DNA methylation analysis. Epigenomics, 13:565-576, 2021
12. Tsuyuki S, Takeshima H, Sekine S, Yamagata Y, Ando T, Yamashita S, Maeda S, Yoshikawa T, Ushijima T. Comparable genetic alteration profiles between gastric cancers with current and past Helicobacter pylori infection. Scientific reports, 11:23443, 2021
13. Hattori N, Asada K, Miyajima N, Mori A, Nakanishi Y, Kimura K, Wakabayashi M, Takeshima H, Nitani C, Hara J, Ushijima T. Combination of a synthetic retinoid and a DNA demethylating agent induced differentiation of neuroblastoma through retinoic acid signal reprogramming. British journal of cancer, 125:1647-1656, 2021
14. Miyamoto T, Uosaki H, Mizunoe Y, Han SI, Goto S, Yamanaka D, Masuda M, Yoneyama Y, Nakamura H, Hattori N, Takeuchi Y, Ohno H, Sekiya M, Matsuzaka T, Hakuno F, Takahashi SI, Yahagi N, Ito K, Shimano H. Rapid manipulation of mitochondrial morphology in a living cell with iCMM. Cell reports methods, 1:100052, 2021
15. Ushijima T, Clark SJ, Tan P. Mapping genomic and epigenomic evolution in cancer ecosystems. Science, 373:1474-1479, 2021
Books
1. García-Giménez JL, Beltrán-García J, Osca-Verdegal R, Pallardó FV, Ushijima T, Tollefsbol TO. Perspectives and future directions of translational epigenetics in personalized and precision medicine. In: García-Giménez JL (ed), Epigenetics in Precision Medicin, Enabled, Academic Press, 2021
