Home > Information > press release > Deep Vision: Near-Infrared Imaging and Machine Learning Can Identify Hidden Tumors
Deep Vision: Near-Infrared Imaging and Machine Learning Can Identify Hidden Tumors
Tokyo, February 2, 2020
in Japanese
Near-infrared hyperspectral imaging combined with machine learning can visualize tumors in deep tissue and covered by a mucosal layer, scientists show
Gastrointestinal stromal tumors are tumors of the digestive tract that grow underneath the mucus layer covering our organs. Because they are deep inside the tissue, these “submucosal tumors” are difficult to detect and diagnose, even with a biopsy. Now, researchers from Japan have developed a novel minimally invasive and accurate method using infrared imaging and machine learning to distinguish between normal tissue and tumor areas. This technique has a strong potential for widespread clinical use.
Tumors can be damaging to surrounding blood vessels and tissues even if they’re benign. If they’re malignant, they’re aggressive and sneaky, and often irrevocably damaging. In the latter case, early detection is key to treatment and recovery. But such detection can sometimes require advanced imaging technology, beyond what is available commercially today.
For instance, some tumors occur deep inside organs and tissues, covered by a mucosal layer, which makes it difficult for scientists to directly observe them with standard methods like endoscopy (which inserts a small camera into a patient’s body via a thin tube) or reach them during biopsies. In particular, gastrointestinal stromal tumors (GISTs)—typically found in the stomach and the small intestines—require demanding techniques that are very time-consuming and prolong the diagnosis. Now, to improve GIST diagnosis, Drs. Daiki Sato, Hiroaki Ikematsu, and Takeshi Kuwata from the National Cancer Center Hospital East in Japan, Dr. Hideo Yokota from the RIKEN Center for Advanced Photonics, Japan, and Drs. Toshihiro Takamatsu and Kohei Soga from Tokyo University of Science, Japan, led by Dr. Hiroshi Takemura, have developed a technology that uses near-infrared hyperspectral imaging (NIR-HSI) along with machine learning. Their findings are published in Nature’s Scientific Reports.
“This technique is a bit like X-rays, the idea is that you use electromagnetic radiation that can pass through the body to generate images of structures inside,” Dr. Takemura explains, “The difference is that X-rays are at 0.01–10 nm, but near-infrared is at around 800–2500 nm. At that wavelength, near-infrared radiation makes tissues seem transparent in images. And these wavelengths are less harmful to the patient than even visible rays.”
This should mean that scientists can safely investigate something that is hidden inside tissues, but until the study by Dr. Takemura and his colleagues, no one had attempted to use NIR-HSI on deep tumors like GISTs. Speaking of what got them to go down this line of investigation, Dr. Takemura pays homage to the late professor who began their journey: “This project has been possible only because of late Prof. Kazuhiro Kaneko, who broke the barriers between doctors and engineers and established this collaboration. We are following his wishes.”
Dr. Takemura’s team performed imaging experiments on 12 patients with confirmed cases of GISTs, who had their tumors removed through surgery. The scientists imaged the excised tissues using NIR-HSI, and then had a pathologist examine the images to determine the border between normal and tumor tissue. These images were then used as training data for a machine-learning algorithm, essentially teaching a computer program to distinguish between the pixels in the images that represent normal tissue versus those that represent tumor tissue.
The scientists found that even though 10 out of the 12 test tumors were completely or partly covered by a mucosal layer, the machine-learning analysis was effective in identifying GISTs, correctly color-coding tumor and non-tumor sections at 86% accuracy. “This is a very exciting development,” Dr. Takemura explains, “Being able to accurately, quickly, and non-invasively diagnose different types of submucosal tumors without biopsies, a procedure that requires surgery, is much easier on both the patient and the physicians.”
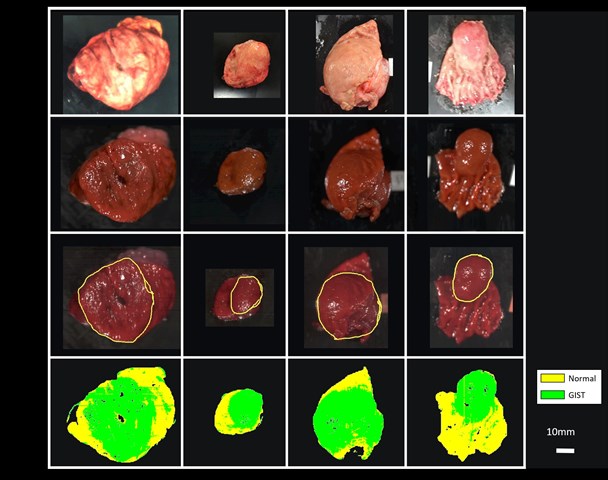
The machine learning technique developed by Dr. Takemura and team could distinguish tumor tissue from healthy tissue in ex vivo images of resected tumors, with 86% accuracy
Photo courtesy: Hiroshi Takemura from Tokyo University of Science
Dr. Takemura acknowledges that there are still challenges ahead, but feels they are prepared to solve them. The researchers identified several areas that would improve on their results, such as making their training dataset much larger, adding information about how deep the tumor is for the machine-learning algorithm, and including other types of tumors in the analysis. Work is also underway to develop an NIR-HSI system that builds on top of existing endoscopy technology.
“We’ve already built a device that attaches an NIR-HSI camera to the end of an endoscope and hope to perform NIR-HSI analysis directly on a patient soon, instead of just on tissues that had been surgically removed,” Dr. Takemura says, “In the future, this will help us separate GISTs from other types of submucosal tumors that could be even more malignant and dangerous. This study is the first step towards much more groundbreaking research in the future, enabled by this interdisciplinary collaboration.”
For now, a means of accurately and non-invasively detecting GISTs early on could be clinically available widely, soon!
Reference
Authors
Daiki Sato1, Toshihiro Takamatsu2,3, Masakazu Umezawa4, Yuichi Kitagawa4, Kosuke Maeda5, Naoki Hosokawa4, Kyohei Okubo4,6, Masao Kamimura4,6, Tomohiro Kadota1, Tetsuo Akimoto7,8, Takahiro Kinoshita9, Tomonori Yano1, Takeshi Kuwata10, Hiroaki Ikematsu1,2, Hiroshi Takemura3,5, Hideo Yokota11 & Kohei Soga3,4,6
Title of original paper
Distinction of surgically resected gastrointestinal stromal tumor by near-infrared hyperspectral imaging
Journal
Scientific Reports
DOI
10.1038/s41598-020-79021-7
Affiliations
1Department of Gastroenterology and Endoscopy, National Cancer Center Hospital East, Japan. 2Exploratory Oncology Research & Clinical Trial Center, National Cancer Center, Japan. 3Research Institute for Biomedical Sciences, Tokyo University of Science, Japan. 4Department of Materials Science and Technology, Tokyo University of Science, Japan. 5Department of Mechanical Engineering, Tokyo University of Science, Japan. 6Imaging Frontier Center, Tokyo University of Science, Japan. 7Division of Radiation Oncology, National Cancer Center Hospital East, Japan. 8Course of Advanced Clinical Research of Cancer, Juntendo University Graduate School of Medicine, Japan. 9Department of Gastric Surgery, National Cancer Center Hospital East, Japan. 10Department of Pathology and Clinical Laboratories, National Cancer Center Hospital East, Japan. 11RIKEN Center for Advanced Photonics, Japan.
Further Information
Professor Hiroshi Takemura
Department of Mechanical Engineering,Tokyo University of Science
Email: takemura@rs.tus.ac.jp
Hiroaki Ikematsu, MD, PhD
Department of Gastroenterology and Endoscopy National Cancer Center Hospital East
Email: hikemats@east.ncc.go.jp
Team Leader: Hideo Yokota (D.Eng.)
Image Processing Research Team RIKEN Center for Advanced Photonics
Email: hyokota@riken.jp
Media contact
Tsutomu Shimizu
Public Relations Divisions Tokyo University of Science
Email: mediaoffice@admin.tus.ac.jp
Website: https://www.tus.ac.jp/en/mediarelations/
Office of Public Relations, Strategic Planning Bureau (Kashiwa Campus) National Cancer Center Japan
E-mail: ncc-admin@ncc.go.jp
Website: https://www.ncc.go.jp/en/index.html
Jens Wilkinson
International Affairs Division RIKEN
Email: gro-pr@riken.jp
Funding information
This study was partially funded by The National Cancer Center Research and Development Fund.
