Home > Organization > Divisions and Independent Research Units > Division of Cellular Signaling > Research Projects
Research Projects
Identification of tumorigenesis
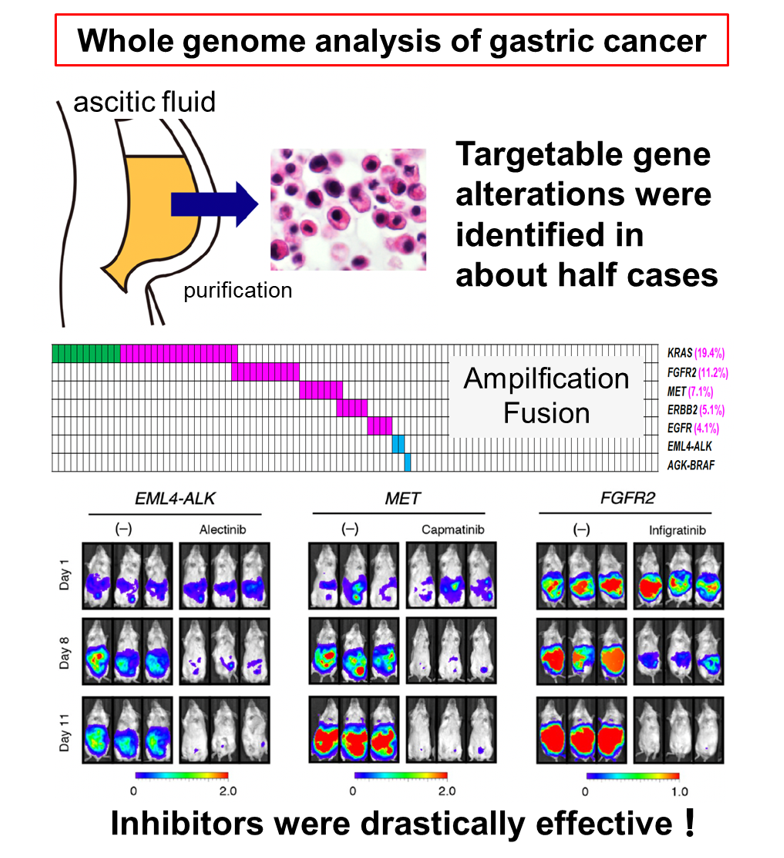
We aim at identifying essential growth drivers in every cancer and developing new molecularly-targeted therapy, by taking advantage of our functional screening system coupled with the next generation sequencing technologies. We recently performed a comprehensive multi-omic analysis of malignant ascitic fluid of advanced gastric cancer and identified potentially targetable gene alterations approximately half of all cases (Nature cancer 2: 962).
Functional assay to evaluate gene alterations
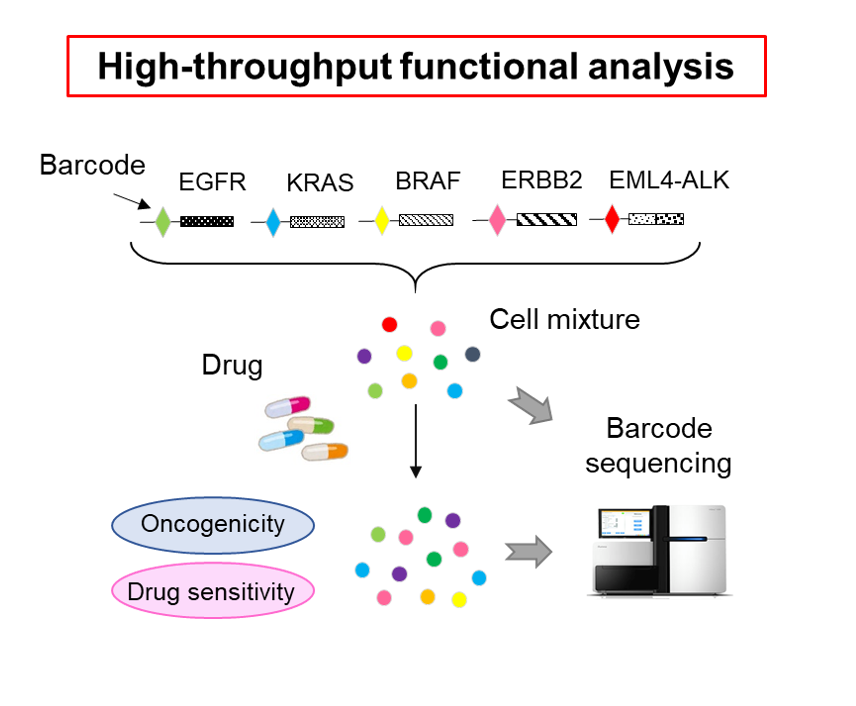
Besides, we are also trying to apply new technologies to the genome medicine. We established a comprehensive assay, the Todai OncoPanel (TOP), which consists of DNA and RNA hybridization capture-based next-generation sequencing panels. A novel method for target enrichment, named the junction capture method, was developed for the RNA panel to accurately and cost-effectively detect most of fusion genes as well as aberrantly spliced transcripts involved oncogenesis in solid tumor. This comprehensive profiling panel is now applied to liquid biopsy using cell-free DNA or circulating tumor cells. Through large-scale cancer genome projects, numerous variants of unknown significance (VUS) have been identified and their functional relevance remains uninvestigated. We developed a high-throughput method to evaluate the transforming potential and drug sensitivity of oncogenes and tumor suppressors and identified novel potential targets in VUS (Sci Transl Med 9:eaan6556,Nat Commun 11: 2573).
Development of new computational pipeline for cancer research
Moreover, we are establishing new computational pipeline for multi-omic analysis by ourselves to optimize for our research purpose. Our motto is“build what we need”while there are many analytical pipeline available these days. For instance, allele sprcific copy number analytical tool (for in-house use) or FFPE specific mutation error elimination tool named MicroSEC (Commun Biol 4: 1396) and fusion detection tool for genome medicine (Cancer Sci 110: 1464) have been developed.

