Home > Organization > Divisions and Independent Research Units > Division of Cellular Signaling > Research Projects > Division of Cellular Signaling
Division of Cellular Signaling
Discovery of EML4-ALK fusion tyrosine kinase, ROS1-fusions and RET-fusions
To identify essential transforming genes from tumor specimens, we had developed in-house a highly sensitive, retroviral cDNA expression library system that can interrogate function of millions of cDNAs isolated even from a very small amount of clinical specimens. By coupling such technology to focus formation assays with fibroblasts, We then started to screen for transforming genes in non-small cell lung cancer (NSCLC), and isolated the first, recurrent fusion-type oncogene, EML4-ALK, in lung cancer (Nature 448:561, 2007).
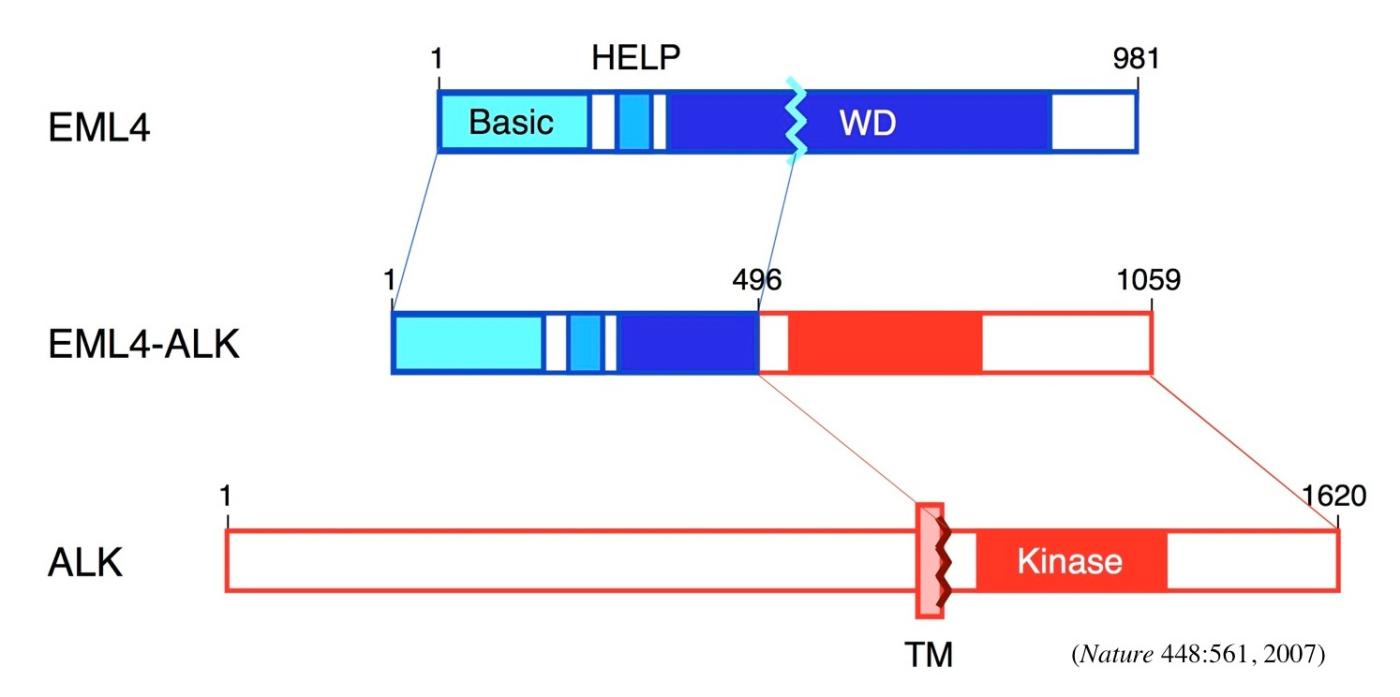
This discovery not only revealed a suitable therapeutic target in highly intractable NSCLC, but also demonstrated one of the earliest evidence for chromosome rearrangement-mediated oncogenesis in major epithelial tumors. Both of EML4 and ALK genes are mapped within the same short arm of human chromosome 2, but a small inversion involving the two loci fuses these genes, and produces a constitutively active and highly oncogenic protein-tyrosine kinase. These findings thus suggest that specific inhibitors against ALK may become “the second Gleevec (targeting BCR-ABL fusion kinase in chronic myeloid leukemia)” in epithelial tumors.
To directly prove EML4-ALK being an ideal therapeutic target in NSCLC, we generated transgenic mice that express the fusion gene specifically in lung alveolar cells. Surprisingly all mice carried hundreds of lung caner nodules at birth, but oral administration of an ALK inhibitor to the mice eradicated such tumors (PNAS 105:19893, 2008). Based on such findings, many pharmaceutical companies raised ALK inhibitors, and one of them, crizotinib, was already approved as a therapeutic drug by U.S. FDA as of August 2011.
We also established RT-PCR–based system to detect EML4-ALK mRNA (Clin Cancer Res 14:6618, 2008), and also a highly sensitive immunohistochemical procedure that reliably detects ELM4-ALK protein among formalin-fixed and paraffin-embedded tissues (Clin Cancer Res 15:3143, 2009), both of which are now under clinical use.
Further, through a genomic analysis of NSLC specimens before and after the relapse against crizotinib treatment, we discovered, for the first time, the secondary mutations within EML4-ALK responsible for the resistance against ALK inhibitors (NEJM 363:1734, 2010).
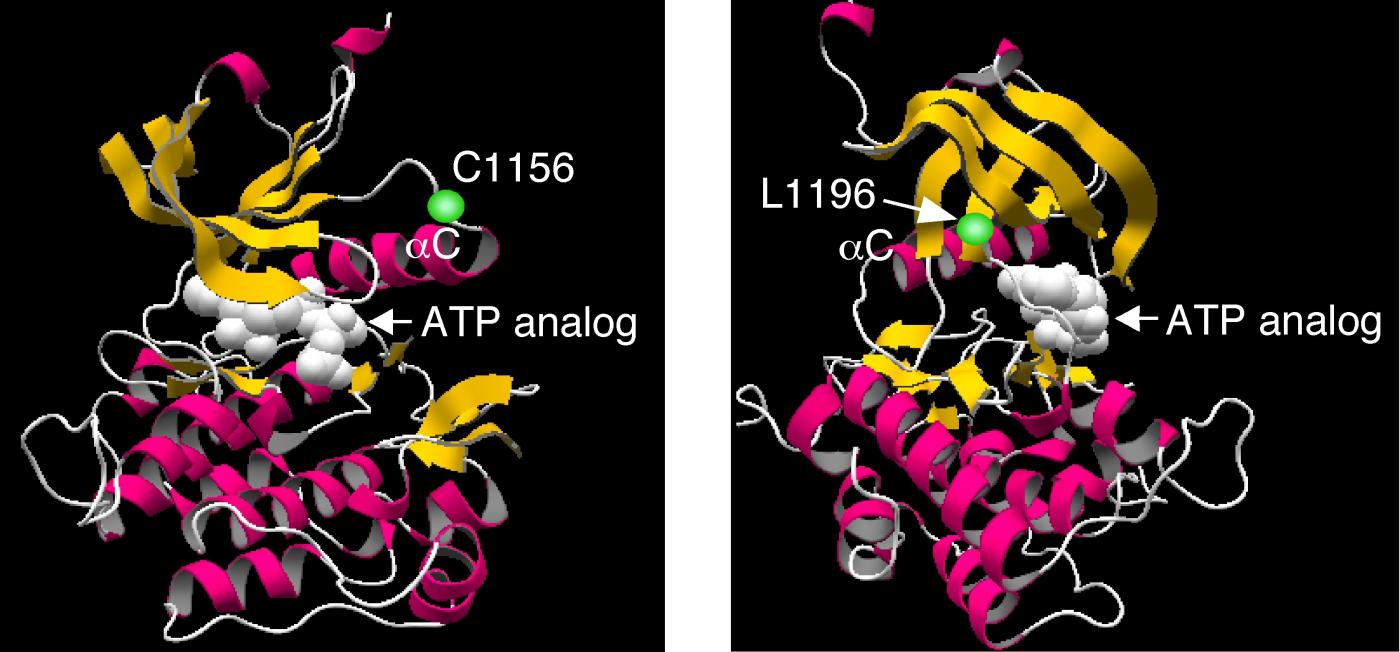
The “second generation” of ALK inhibitors have been developed based on this discovery, and six of them are already under clinical trials. The outcome of a phase I/II trial for one of such ALK inhibitors, CH542802, was recently reported, showing a magical response rate of 93.5% as a single drug treatment against NSCLC (Lancet Oncol 14:590, 2013).
Our activities thus newly generated and led a large translational research field of “Targeting ALK in epithelial tumors”, by the discovery of EML4-ALK in lung cancer, development of the diagnostic scheme through our volunteer initiative, and the identification of the mechanism for the acquired resistance. Furthermore, we subsequently discovered activating point mutations within ALK among individuals with neuroblastoma (Nature 455:971, 2008) and RET fusions-and ROS1-fusions in NSCLC (Nat Med 18:378, 2012).
Cancer genome resequencing with the next-generation sequencers
The next-generation sequencer can generate 300-600 Gbp of sequence data in a single experiment. To efficiently identify cancer-causing genes, we thus combine (i) the mutation-search for cancer genome with the next-generation sequencer and (ii) the sensitive functional screening system that succeeded in the discovery of EML4-ALK.
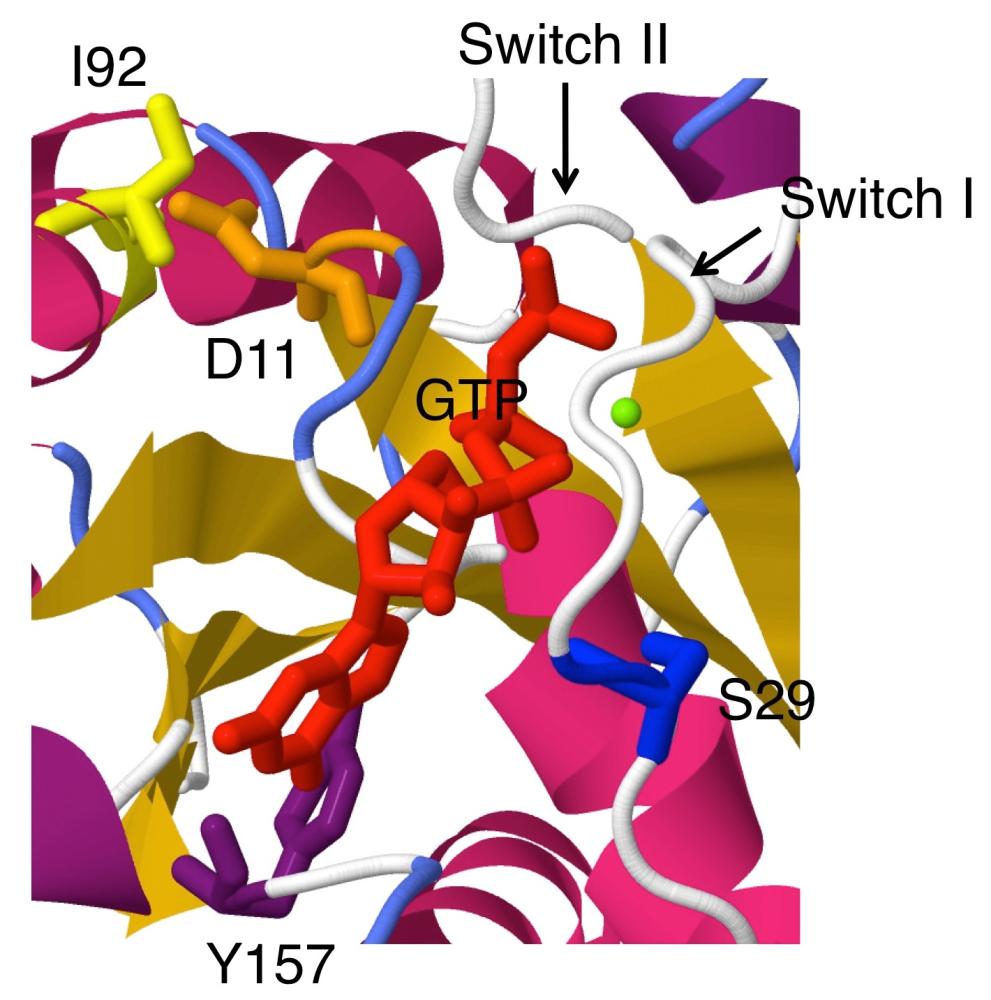
Application of this approach to a human sarcoma cell line HT1080 led to identify the N92I mutant of a small G-protein RAC1 (PNAS 10:3029, 2013). RAC1(N92I) is a potent oncoprotein, and this cell line is dependent on RAC1(N92I) for proliferation. Screening of other mutations in RAC family isolated oncogenic RAC1(P29S), RAC1(C157Y) and RAC2(P29Q). Thus, RAC-family mutants are potential therapeutic targets.